Fast hydrogen exchange affects ¹⁵N relaxation measurements in intrinsically disordered proteins
- PMID: 23314729
- PMCID: PMC3615062
- DOI: 10.1007/s10858-013-9706-1
Fast hydrogen exchange affects ¹⁵N relaxation measurements in intrinsically disordered proteins
Abstract
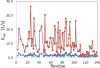
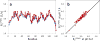
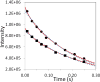
Unprotected amide protons can undergo fast hydrogen exchange (HX) with protons from the solvent. Generally, NMR experiments using the out-and-back coherence transfer with amide proton detection are affected by fast HX and result in reduced signal intensity. When one of these experiments, (1)H-(15)N HSQC, is used to measure the (15)N transverse relaxation rate (R2), the measured R2 rate is convoluted with the HX rate (kHX) and has higher apparent R2 values. Since the (15)N R2 measurement is important for analyzing protein backbone dynamics, the HX effect on the R2 measurement is investigated and described here by multi-exponential signal decay. We demonstrate these effects by performing (15)N R 2 (CPMG) experiments on α-synuclein, an intrinsically disordered protein, in which the amide protons are exposed to solvent. We show that the HX effect on R 2 (CPMG) can be extracted by the derived equation. In conclusion, the HX effect may be pulse sequence specific and results from various sources including the J coupling evolution, the change of steady state water proton magnetization, and the D2O content in the sample. To avoid the HX effect on the analysis of relaxation data of unprotected amides, it is suggested that NMR experimental conditions insensitive to the HX should be considered or that intrinsic R 2 (CPMG) values be obtained by methods described herein.
Figures

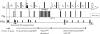
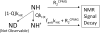
References
-
- Bertini I, Carrano CJ, Luchinat C, Piccioli M, Poggi L. A 15N NMR mobility study on the dicalcium P43M calbindin D9k and its mono-La3+-substituted form. Biochemistry. 2002;41:5104–5111. - PubMed
-
- Bertoncini CW, Rasia RM, Lamberto GR, Binolfi A, Zweckstetter M, Griesinger C, Fernandez CO. Structural characterization of the intrinsically unfolded protein beta-synuclein, a natural negative regulator of alpha-synuclein aggregation. J Mol Biol. 2007;372:708–722. - PubMed
-
- Buevich AV, Baum J. Dynamics of unfolded proteins: incorporation of distributions of correlation times in the model free analysis of NMR relaxation data. J Am Chem Soc. 1999;121:8671–8672.
-
- Buevich AV, Shinde UP, Inouye M, Baum J. Backbone dynamics of the natively unfolded pro-peptide of subtilisin by heteronuclear NMR relaxation studies. J Biomol NMR. 2001;20:233–249. - PubMed
-
- Bussell R, Jr, Eliezer D. Residual structure and dynamics in Parkinson’s disease-associated mutants of alpha-synuclein. J Biol Chem. 2001;276:45996–46003. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

