Carbon and nitrogen provisions alter the metabolic flux in developing soybean embryos
- PMID: 23314943
- PMCID: PMC3585609
- DOI: 10.1104/pp.112.203299
Carbon and nitrogen provisions alter the metabolic flux in developing soybean embryos
Abstract
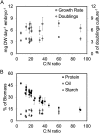
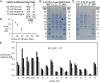
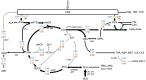
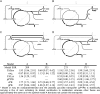
Soybean (Glycine max) seeds store significant amounts of their biomass as protein, levels of which reflect the carbon and nitrogen received by the developing embryo. The relationship between carbon and nitrogen supply during filling and seed composition was examined through a series of embryo-culturing experiments. Three distinct ratios of carbon to nitrogen supply were further explored through metabolic flux analysis. Labeling experiments utilizing [U-(13)C5]glutamine, [U-(13)C4]asparagine, and [1,2-(13)C2]glucose were performed to assess embryo metabolism under altered feeding conditions and to create corresponding flux maps. Additionally, [U-(14)C12]sucrose, [U-(14)C6]glucose, [U-(14)C5]glutamine, and [U-(14)C4]asparagine were used to monitor differences in carbon allocation. The analyses revealed that: (1) protein concentration as a percentage of total soybean embryo biomass coincided with the carbon-to-nitrogen ratio; (2) altered nitrogen supply did not dramatically impact relative amino acid or storage protein subunit profiles; and (3) glutamine supply contributed 10% to 23% of the carbon for biomass production, including 9% to 19% of carbon to fatty acid biosynthesis and 32% to 46% of carbon to amino acids. Seed metabolism accommodated different levels of protein biosynthesis while maintaining a consistent rate of dry weight accumulation. Flux through ATP-citrate lyase, combined with malic enzyme activity, contributed significantly to acetyl-coenzyme A production. These fluxes changed with plastidic pyruvate kinase to maintain a supply of pyruvate for amino and fatty acids. The flux maps were independently validated by nitrogen balancing and highlight the robustness of primary metabolism.
Figures




References
-
- Adams CA, Broman TH, Rinne RW. (1982) Use of [3,4-C]glucose to assess in vivo competition for phosphoenolpyruvate between phosphoenolpyruvate carboxylase and pyruvate kinase in developing soybean seeds. Plant Cell Physiol 23: 959–965
-
- Adams CA, Rinne RW. (1981) Interactions of phosphoenolpyruvate carboxylase and pyruvic kinase in developing soybean seeds. Plant Cell Physiol 22: 1011–1021
-
- Allen DK, Libourel IGL, Shachar-Hill Y. (2009a) Metabolic flux analysis in plants: coping with complexity. Plant Cell Environ 32: 1241–1257 - PubMed
-
- Allen DK, Ohlrogge JB, Shachar-Hill Y. (2009b) The role of light in soybean seed filling metabolism. Plant J 58: 220–234 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

