Bcr1 functions downstream of Ssd1 to mediate antimicrobial peptide resistance in Candida albicans
- PMID: 23314964
- PMCID: PMC3629773
- DOI: 10.1128/EC.00285-12
Bcr1 functions downstream of Ssd1 to mediate antimicrobial peptide resistance in Candida albicans
Abstract
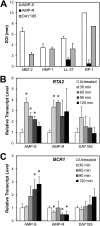
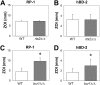
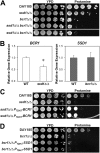
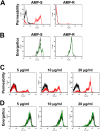
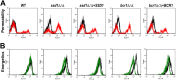
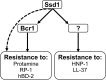
In order to colonize the host and cause disease, Candida albicans must avoid being killed by host defense peptides. Previously, we determined that the regulatory protein Ssd1 governs antimicrobial peptide resistance in C. albicans. Here, we sought to identify additional genes whose products govern susceptibility to antimicrobial peptides. We discovered that a bcr1Δ/Δ mutant, like the ssd1Δ/Δ mutant, had increased susceptibility to the antimicrobial peptides, protamine, RP-1, and human β defensin-2. Homozygous deletion of BCR1 in the ssd1Δ/Δ mutant did not result in a further increase in antimicrobial peptide susceptibility. Exposure of the bcr1Δ/Δ and ssd1Δ/Δ mutants to RP-1 induced greater loss of mitochondrial membrane potential and increased plasma membrane permeability than with the control strains. Therefore, Bcr1 and Ssd1 govern antimicrobial peptide susceptibility and likely function in the same pathway. Furthermore, BCR1 mRNA expression was downregulated in the ssd1Δ/Δ mutant, and the forced expression of BCR1 in the ssd1Δ/Δ mutant partially restored antimicrobial peptide resistance. These results suggest that Bcr1 functions downstream of Ssd1. Interestingly, overexpression of 11 known Bcr1 target genes in the bcr1Δ/Δ mutant failed to restore antimicrobial peptide resistance, suggesting that other Bcr1 target genes are likely responsible for antimicrobial peptide resistance. Collectively, these results demonstrate that Bcr1 functions downstream of Ssd1 to govern antimicrobial peptide resistance by maintaining mitochondrial energetics and reducing membrane permeabilization.
Figures


References
-
- Yount NY, Yeaman MR. 2012. Emerging themes and therapeutic prospects for anti-infective peptides. Annu. Rev. Pharmacol. Toxicol. 52: 337– 360 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

