Human immunodeficiency viruses appear compartmentalized to the female genital tract in cross-sectional analyses but genital lineages do not persist over time
- PMID: 23315326
- PMCID: PMC3603533
- DOI: 10.1093/infdis/jit016
Human immunodeficiency viruses appear compartmentalized to the female genital tract in cross-sectional analyses but genital lineages do not persist over time
Abstract
Background: Whether unique human immunodeficiency type 1 (HIV) genotypes occur in the genital tract is important for vaccine development and management of drug resistant viruses. Multiple cross-sectional studies suggest HIV is compartmentalized within the female genital tract. We hypothesize that bursts of HIV replication and/or proliferation of infected cells captured in cross-sectional analyses drive compartmentalization but over time genital-specific viral lineages do not form; rather viruses mix between genital tract and blood.
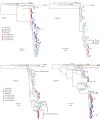
Methods: Eight women with ongoing HIV replication were studied during a period of 1.5 to 4.5 years. Multiple viral sequences were derived by single-genome amplification of the HIV C2-V5 region of env from genital secretions and blood plasma. Maximum likelihood phylogenies were evaluated for compartmentalization using 4 statistical tests.
Results: In cross-sectional analyses compartmentalization of genital from blood viruses was detected in three of eight women by all tests; this was associated with tissue specific clades containing multiple monotypic sequences. In longitudinal analysis, the tissues-specific clades did not persist to form viral lineages. Rather, across women, HIV lineages were comprised of both genital tract and blood sequences.
Conclusions: The observation of genital-specific HIV clades only in cross-sectional analysis and an absence of genital-specific lineages in longitudinal analyses suggest a dynamic interchange of HIV variants between the female genital tract and blood.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

