Salmonella and Escherichia coli O157:H7 survival in soil and translocation into leeks (Allium porrum) as influenced by an arbuscular mycorrhizal fungus (Glomus intraradices)
- PMID: 23315740
- PMCID: PMC3592219
- DOI: 10.1128/AEM.02855-12
Salmonella and Escherichia coli O157:H7 survival in soil and translocation into leeks (Allium porrum) as influenced by an arbuscular mycorrhizal fungus (Glomus intraradices)
Abstract
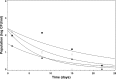
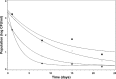
A study was conducted to determine the influence of arbuscular mycorrhizal (AM) fungi on Salmonella and enterohemorrhagic Escherichia coli O157:H7 (EHEC) in autoclaved soil and translocation into leek plants. Six-week-old leek plants (with [Myc+] or without [Myc-] AM fungi) were inoculated with composite suspensions of Salmonella or EHEC at ca. 8.2 log CFU/plant into soil. Soil, root, and shoot samples were analyzed for pathogens on days 1, 8, 15, and 22 postinoculation. Initial populations (day 1) were ca. 3.1 and 2.1 log CFU/root, ca. 2.0 and 1.5 log CFU/shoot, and ca. 5.5 and 5.1 CFU/g of soil for Salmonella and EHEC, respectively. Enrichments indicated that at days 8 and 22, only 31% of root samples were positive for EHEC, versus 73% positive for Salmonella. The mean Salmonella level in soil was 3.4 log CFU/g at day 22, while EHEC populations dropped to ≤ 0.75 log CFU/g by day 15. Overall, Salmonella survived in a greater number of shoot, root, and soil samples, compared with the survival of EHEC. EHEC was not present in Myc- shoots after day 8 (0/16 samples positive); however, EHEC persisted in higher numbers (P = 0.05) in Myc+ shoots (4/16 positive) at days 15 and 22. Salmonella, likewise, survived in statistically higher numbers of Myc+ shoot samples (8/8) at day 8, compared with survival in Myc- shoots (i.e., only 4/8). These results suggest that AM fungi may potentially enhance the survival of E. coli O157:H7 and Salmonella in the stems of growing leek plants.
Figures


Similar articles
-
Influence of mycorrhizal fungi on fate of E. coli O157:H7 and Salmonella in soil and internalization into Romaine lettuce plants.Int J Food Microbiol. 2015 Jan 2;192:95-102. doi: 10.1016/j.ijfoodmicro.2014.10.001. Epub 2014 Oct 7. Int J Food Microbiol. 2015. PMID: 25440552
-
Persistence of enterohaemorrhagic and nonpathogenic E. coli on spinach leaves and in rhizosphere soil.J Appl Microbiol. 2010 May;108(5):1789-96. doi: 10.1111/j.1365-2672.2009.04583.x. Epub 2009 Oct 12. J Appl Microbiol. 2010. PMID: 19878527
-
Fate of Escherichia coli O157:H7 in manure compost-amended soil and on carrots and onions grown in an environmentally controlled growth chamber.J Food Prot. 2004 Mar;67(3):574-8. doi: 10.4315/0362-028x-67.3.574. J Food Prot. 2004. PMID: 15035376
-
Fate of Salmonella enterica and Enterohemorrhagic Escherichia coli on Vegetable Seeds Contaminated by Direct Contact with Artificially Inoculated Soil during Germination.J Food Prot. 2020 Jul 1;83(7):1218-1226. doi: 10.4315/JFP-20-021. J Food Prot. 2020. PMID: 32221551
-
Adherence factors of enterohemorrhagic Escherichia coli O157:H7 strain Sakai influence its uptake into the roots of Valerianella locusta grown in soil.Food Microbiol. 2018 Dec;76:245-256. doi: 10.1016/j.fm.2018.05.016. Epub 2018 May 31. Food Microbiol. 2018. PMID: 30166148
Cited by
-
Spread and change in stress resistance of Shiga toxin-producing Escherichia coli O157 on fungal colonies.Microb Biotechnol. 2014 Nov;7(6):621-9. doi: 10.1111/1751-7915.12071. Epub 2013 Aug 6. Microb Biotechnol. 2014. PMID: 23919289 Free PMC article.
References
-
- Bernstein N, Sela S, Pinto R, Loffe M. 2007. Evidence for internalization of Escherichia coli into the aerial parts of maize via the root system. J. Food Prot. 70:471–475 - PubMed
-
- Deering AJ, Pruitt RE, Mauer LJ, Reuhs BL. 2012. Examination of the internalization of Salmonella serovar Typhimurium in peanut, Arachis hypogaea, using immunocytochemical techniques. Food Res. Int. 45:1037–1043 http://www.sciencedirect.com/science/article/pii/S0963996911000846
-
- Franz E, Visser AA, Van Diepeningen AD, Klerks M, Termorshuizen AJ, van Bruggen AHC. 2007. Quantification of contamination of lettuce by GFP-expressing Escherichia coli O157 and Salmonella enterica serovar Typhimurium. Food Microbiol. 24:106–112 - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

