Examination of food chain-derived Listeria monocytogenes strains of different serotypes reveals considerable diversity in inlA genotypes, mutability, and adaptation to cold temperatures
- PMID: 23315746
- PMCID: PMC3592241
- DOI: 10.1128/AEM.03341-12
Examination of food chain-derived Listeria monocytogenes strains of different serotypes reveals considerable diversity in inlA genotypes, mutability, and adaptation to cold temperatures
Abstract
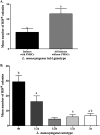
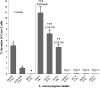
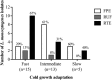
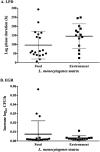
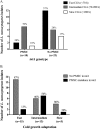
Listeria monocytogenes strains belonging to serotypes 1/2a and 4b are frequently linked to listeriosis. While inlA mutations leading to premature stop codons (PMSCs) and attenuated virulence are common in 1/2a, they are rare in serotype 4b. We observed PMSCs in 35% of L. monocytogenes isolates (n = 54) recovered from the British Columbia food supply, including serotypes 1/2a (30%), 1/2c (100%), and 3a (100%), and a 3-codon deletion (amino acid positions 738 to 740) seen in 57% of 4b isolates from fish-processing facilities. Caco-2 invasion assays showed that two isolates with the deletion were significantly more invasive than EGD-SmR (P < 0.0001) and were either as (FF19-1) or more (FE13-1) invasive than a clinical control strain (08-5578) (P = 0.006). To examine whether serotype 1/2a was more likely to acquire mutations than other serotypes, strains were plated on agar with rifampin, revealing 4b isolates to be significantly more mutable than 1/2a, 1/2c, and 3a serotypes (P = 0.0002). We also examined the ability of 33 strains to adapt to cold temperature following a downshift from 37°C to 4°C. Overall, three distinct cold-adapting groups (CAG) were observed: 46% were fast (<70 h), 39% were intermediate (70 to 200 h), and 15% were slow (>200 h) adaptors. Intermediate CAG strains (70%) more frequently possessed inlA PMSCs than did fast (20%) and slow (10%) CAGs; in contrast, 87% of fast adaptors lacked inlA PMSCs. In conclusion, we report food chain-derived 1/2a and 4b serotypes with a 3-codon deletion possessing invasive behavior and the novel association of inlA genotypes encoding a full-length InlA with fast cold-adaptation phenotypes.
Figures
References
-
- Swaminathan B, Gerner-Smidt P. 2007. The epidemiology of human listeriosis. Microbes Infect. 9:1236–1243 - PubMed
-
- McLauchlin J, Mitchell RT, Smerdon WJ, Jewell K. 2004. Listeria monocytogenes and listeriosis: a review of hazard characterisation for use in microbiological risk assessment of foods. Int. J. Food Microbiol. 92:15–33 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

