Protein lysine-Nζ alkylation and O-phosphorylation mediated by DTT-generated reactive oxygen species
- PMID: 23315912
- PMCID: PMC3595463
- DOI: 10.1002/pro.2214
Protein lysine-Nζ alkylation and O-phosphorylation mediated by DTT-generated reactive oxygen species
Abstract
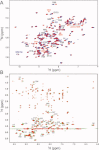
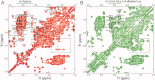
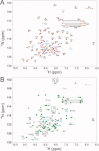
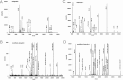
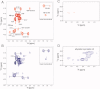
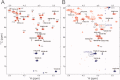
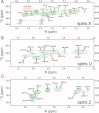
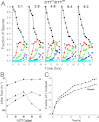
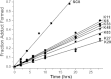
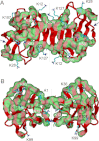
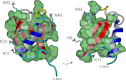
Reactive oxygen species (ROS) play crucial roles in physiology and pathology. In this report, we use NMR spectroscopy and mass spectrometry (MS) to demonstrate that proteins (galectin-1, ubiquitin, RNase, cytochrome c, myoglobin, and lysozyme) under reducing conditions with dithiothreitol (DTT) become alkylated at lysine-N(ζ) groups and O-phosphorylated at serine and threonine residues. These adduction reactions only occur in the presence of monophosphate, potassium, trace metals Fe/Cu, and oxygen, and are promoted by reactive oxygen species (ROS) generated via DTT oxidation. Superoxide mediates the chemistry, because superoxide dismutase inhibits the reaction, and hydroxyl and phosphoryl radicals are also likely involved. While lysine alkylation accounts for most of the adduction, low levels of phosphorylation are also observed at some serine and threonine residues, as determined by western blotting and MS fingerprinting. The adducted alkyl group is found to be a fragment of DTT that forms a Schiff base at lysine N(ζ) groups. Although its exact chemical structure remains unknown, the DTT fragment includes a SH group and a --CHOH--CH₂-- group. Chemical adduction appears to be promoted in the context of a well-folded protein, because some adducted sites in the proteins studied are considerably more reactive than others and the reaction occurs to a lesser extent with shorter, unfolded peptides and not at all with small organic molecules. A structural signature involving clusters of positively charged and other polar groups appears to facilitate the reaction. Overall, our findings demonstrate a novel reaction for DTT-mediated ROS chemistry with proteins.
Copyright © 2013 The Protein Society.
Figures


Similar articles
-
Rethinking Dithiothreitol-Based Particulate Matter Oxidative Potential: Measuring Dithiothreitol Consumption versus Reactive Oxygen Species Generation.Environ Sci Technol. 2017 Jun 6;51(11):6507-6514. doi: 10.1021/acs.est.7b01272. Epub 2017 May 18. Environ Sci Technol. 2017. PMID: 28489384
-
Quantitative analysis of both protein expression and serine / threonine post-translational modifications through stable isotope labeling with dithiothreitol.Proteomics. 2005 Feb;5(2):388-98. doi: 10.1002/pmic.200401066. Proteomics. 2005. PMID: 15648052
-
Identification of an acetonitrile addition impurity formed during peptide disulfide bond reduction using dithiothreitol and Tris(2-carboxyethyl)phosphine.J Pharm Biomed Anal. 2019 Sep 10;174:518-524. doi: 10.1016/j.jpba.2019.06.020. Epub 2019 Jun 19. J Pharm Biomed Anal. 2019. PMID: 31252308
-
[Advancesin enrichment and detection methods for N-phosphorylated proteins].Se Pu. 2020 Mar 8;38(3):278-286. doi: 10.3724/SP.J.1123.2019.09015. Se Pu. 2020. PMID: 34213207 Review. Chinese.
-
Interconnection of reactive oxygen species chemistry across the interfaces of atmospheric, environmental, and biological processes.Acc Chem Res. 2015 Mar 17;48(3):575-83. doi: 10.1021/ar500412p. Epub 2015 Feb 17. Acc Chem Res. 2015. PMID: 25688469 Review.
Cited by
-
Distinct Roles of Nrf1 and Nrf2 in Monitoring the Reductive Stress Response to Dithiothreitol (DTT).Antioxidants (Basel). 2022 Aug 7;11(8):1535. doi: 10.3390/antiox11081535. Antioxidants (Basel). 2022. PMID: 36009254 Free PMC article.
References
-
- Misra HP. Generation of superoxide free radical during the autoxidation of thiols. J Biol Chem. 1974;249:2151–2155. - PubMed
-
- Kachur AV, Held KD, Koch CJ, Biaglow JE. Mechanism of production of hydroxyl radicals in the copper-catalyzed oxidation of dithiothreitol. Radiat Res. 1997;147:409–415. - PubMed
-
- Devasagayam TP, Tilak JC, Boloor KK, Sane KS, Ghaskadbi SS, Lele RD. Free radicals and antioxidants in human health: current status and future prospects. J Assoc Phys India. 2004;52:794–804. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

