Combination of culture-independent and culture-dependent molecular methods for the determination of bacterial community of iru, a fermented Parkia biglobosa seeds
- PMID: 23316189
- PMCID: PMC3539807
- DOI: 10.3389/fmicb.2012.00436
Combination of culture-independent and culture-dependent molecular methods for the determination of bacterial community of iru, a fermented Parkia biglobosa seeds
Abstract
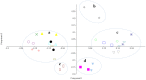
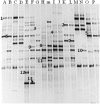
In this study, bacterial composition of iru produced by natural, uncontrolled fermentation of Parkia biglobosa seeds was assessed using culture-independent method in combination with culture-based genotypic typing techniques. PCR-denaturing gradient gel electrophoresis (DGGE) revealed similarity in DNA fragments with the two DNA extraction methods used and confirmed bacterial diversity in the 16 iru samples from different production regions. DNA sequencing of the highly variable V3 region of the 16S rRNA genes obtained from PCR-DGGE identified species related to Bacillus subtilis as consistent bacterial species in the fermented samples, while other major bands were identified as close relatives of Staphylococcus vitulinus, Morganella morganii, B. thuringiensis, S. saprophyticus, Tetragenococcus halophilus, Ureibacillus thermosphaericus, Brevibacillus parabrevis, Salinicoccus jeotgali, Brevibacterium sp. and uncultured bacteria clones. Bacillus species were cultured as potential starter cultures and clonal relationship of different isolates determined using amplified ribosomal DNA restriction analysis (ARDRA) combined with 16S-23S rRNA gene internal transcribed spacer (ITS) PCR amplification, restriction analysis (ITS-PCR-RFLP), and randomly amplified polymorphic DNA (RAPD-PCR). This further discriminated B. subtilis and its variants from food-borne pathogens such as B. cereus and suggested the need for development of controlled fermentation processes and good manufacturing practices (GMP) for iru production to achieve product consistency, safety quality, and improved shelf life.
Keywords: Bacillus; DGGE; fermentation; iru; safety.
Figures
References
-
- Abriouel H., Ben Omar N., López R. L., Cañamero M. M., Ortega E., Gálvez A. (2007). Differentiation and characterization by molecular techniques of Bacillus cereus group isolates from poto poto and degue, two traditional cereal-based fermented foods of Burkina Faso and Republic of Congo. J. Food Prot. 70, 1165–1173 - PubMed
-
- Achi O. K. (2005). Traditional fermented protein condiments in Nigeria. Afr. J. Biotechnol. 4, 1612–1621
-
- Ampe F., Miambi E. (2000). Cluster analysis, richness and biodiversity indexes derived from denaturing gradient gel electrophoresis fingerprints of bacterial communities demonstrate that traditional maize fermentations are driven by transformation process. Int. J. Food Microbiol. 60, 91–97 10.1016/S0168-1605(00)00358-5 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

