Genetic analysis of mlh3 mutations reveals interactions between crossover promoting factors during meiosis in baker's yeast
- PMID: 23316435
- PMCID: PMC3538346
- DOI: 10.1534/g3.112.004622
Genetic analysis of mlh3 mutations reveals interactions between crossover promoting factors during meiosis in baker's yeast
Abstract
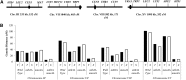
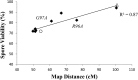
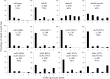
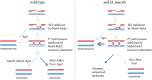
Crossing over between homologous chromosomes occurs during the prophase of meiosis I and is critical for chromosome segregation. In baker's yeast, two heterodimeric complexes, Msh4-Msh5 and Mlh1-Mlh3, act in meiosis to promote interference-dependent crossing over. Mlh1-Mlh3 also plays a role in DNA mismatch repair (MMR) by interacting with Msh2-Msh3 to repair insertion and deletion mutations. Mlh3 contains an ATP-binding domain that is highly conserved among MLH proteins. To explore roles for Mlh3 in meiosis and MMR, we performed a structure-function analysis of eight mlh3 ATPase mutants. In contrast to previous work, our data suggest that ATP hydrolysis by both Mlh1 and Mlh3 is important for both meiotic and MMR functions. In meiotic assays, these mutants showed a roughly linear relationship between spore viability and genetic map distance. To further understand the relationship between crossing over and meiotic viability, we analyzed crossing over on four chromosomes of varying lengths in mlh3Δ mms4Δ strains and observed strong decreases (6- to 17-fold) in crossing over in all intervals. Curiously, mlh3Δ mms4Δ double mutants displayed spore viability levels that were greater than observed in mms4Δ strains that show modest defects in crossing over. The viability in double mutants also appeared greater than would be expected for strains that show such severe defects in crossing over. Together, these observations provide insights for how Mlh1-Mlh3 acts in crossover resolution and MMR and for how chromosome segregation in Meiosis I can occur in the absence of crossing over.
Keywords: DNA mismatch repair; Mlh1-Mlh3; Msh4-Msh5; crossing over; meiotic recombination.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
