Enhanced ranking of PknB Inhibitors using data fusion methods
- PMID: 23317154
- PMCID: PMC3600029
- DOI: 10.1186/1758-2946-5-2
Enhanced ranking of PknB Inhibitors using data fusion methods
Abstract
Background: Mycobacterium tuberculosis encodes 11 putative serine-threonine proteins Kinases (STPK) which regulates transcription, cell development and interaction with the host cells. From the 11 STPKs three kinases namely PknA, PknB and PknG have been related to the mycobacterial growth. From previous studies it has been observed that PknB is essential for mycobacterial growth and expressed during log phase of the growth and phosphorylates substrates involved in peptidoglycan biosynthesis. In recent years many high affinity inhibitors are reported for PknB. Previously implementation of data fusion has shown effective enrichment of active compounds in both structure and ligand based approaches .In this study we have used three types of data fusion ranking algorithms on the PknB dataset namely, sum rank, sum score and reciprocal rank. We have identified reciprocal rank algorithm is capable enough to select compounds earlier in a virtual screening process. We have also screened the Asinex database with reciprocal rank algorithm to identify possible inhibitors for PknB.
Results: In our work we have used both structure-based and ligand-based approaches for virtual screening, and have combined their results using a variety of data fusion methods. We found that data fusion increases the chance of actives being ranked highly. Specifically, we found that the ranking of Pharmacophore search, ROCS and Glide XP fused with a reciprocal ranking algorithm not only outperforms structure and ligand based approaches but also capable of ranking actives better than the other two data fusion methods using the BEDROC, robust initial enhancement (RIE) and AUC metrics. These fused results were used to identify 45 candidate compounds for further experimental validation.
Conclusion: We show that very different structure and ligand based methods for predicting drug-target interactions can be combined effectively using data fusion, outperforming any single method in ranking of actives. Such fused results show promise for a coherent selection of candidates for biological screening.
Figures
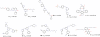
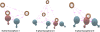

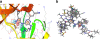

References
LinkOut - more resources
Full Text Sources
Other Literature Sources

