UniChem: a unified chemical structure cross-referencing and identifier tracking system
- PMID: 23317286
- PMCID: PMC3616875
- DOI: 10.1186/1758-2946-5-3
UniChem: a unified chemical structure cross-referencing and identifier tracking system
Abstract
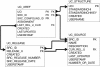
UniChem is a freely available compound identifier mapping service on the internet, designed to optimize the efficiency with which structure-based hyperlinks may be built and maintained between chemistry-based resources. In the past, the creation and maintenance of such links at EMBL-EBI, where several chemistry-based resources exist, has required independent efforts by each of the separate teams. These efforts were complicated by the different data models, release schedules, and differing business rules for compound normalization and identifier nomenclature that exist across the organization. UniChem, a large-scale, non-redundant database of Standard InChIs with pointers between these structures and chemical identifiers from all the separate chemistry resources, was developed as a means of efficiently sharing the maintenance overhead of creating these links. Thus, for each source represented in UniChem, all links to and from all other sources are automatically calculated and immediately available for all to use. Updated mappings are immediately available upon loading of new data releases from the sources. Web services in UniChem provide users with a single simple automatable mechanism for maintaining all links from their resource to all other sources represented in UniChem. In addition, functionality to track changes in identifier usage allows users to monitor which identifiers are current, and which are obsolete. Lastly, UniChem has been deliberately designed to allow additional resources to be included with minimal effort. Indeed, the recent inclusion of data sources external to EMBL-EBI has provided a simple means of providing users with an even wider selection of resources with which to link to, all at no extra cost, while at the same time providing a simple mechanism for external resources to link to all EMBL-EBI chemistry resources.
Figures


References
-
- ChEBI. http://www.ebi.ac.uk/chebi.
-
- ChEMBL. https://www.ebi.ac.uk/chembl.
-
- PDBe. http://www.ebi.ac.uk/pdbe.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

