The piRNA pathway in flies: highlights and future directions
- PMID: 23317515
- PMCID: PMC3621807
- DOI: 10.1016/j.gde.2012.12.003
The piRNA pathway in flies: highlights and future directions
Abstract
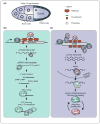
Piwi proteins, together with their bound Piwi-interacting RNAs, constitute an evolutionarily conserved, germline-specific innate immune system. The piRNA pathway is one of the key mechanisms for silencing transposable elements in the germline, thereby preserving genome integrity between generations. Recent work from several groups has significantly advanced our understanding of how piRNAs arise from discrete genomic loci, termed piRNA clusters, and how these Piwi-piRNA complexes enforce transposon silencing. Here, we discuss these recent findings, as well as highlight some aspects of piRNA biology that continue to escape our understanding.
Copyright © 2013. Published by Elsevier Ltd.
Figures

References
-
- Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol. 2011;12:246–258. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

