Induction of cells with cancer stem cell properties from nontumorigenic human mammary epithelial cells by defined reprogramming factors
- PMID: 23318426
- PMCID: PMC4697746
- DOI: 10.1038/onc.2012.614
Induction of cells with cancer stem cell properties from nontumorigenic human mammary epithelial cells by defined reprogramming factors
Abstract
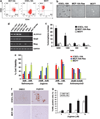
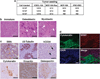
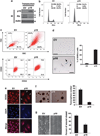
Cancer stem cells (CSCs), a small and elusive population of undifferentiated cancer cells within tumors that drive tumor growth and recurrence, are believed to resemble normal stem cells. Although surrogate markers have been identified and compelling CSC theoretical models abound, actual proof for the existence of CSCs can only be had retrospectively. Hence, great store has come to be placed in isolating CSCs from cancers for in-depth analysis. On the other hand, although induced pluripotent stem cells (iPSCs) hold great promise for regenerative medicine, concern exists over the inadvertent co-transplantation of partially or undifferentiated stem cells with tumorigenic capacity. Here we demonstrate that the introduction of defined reprogramming factors (OCT4, SOX2, Klf4 and c-Myc) into MCF-10A nontumorigenic mammary epithelial cells, followed by partial differentiation, transforms the bulk of cells into tumorigenic CD44(+)/CD24(low) cells with CSC properties, termed here as induced CSC-like-10A or iCSCL-10A cells. These reprogrammed cells display a malignant phenotype in culture and form tumors of multiple lineages when injected into immunocompromised mice. Compared with other transformed cell lines, cultured iCSCL-10A cells exhibit increased resistance to the chemotherapeutic compounds, Taxol and Actinomycin D, but higher susceptibility to the CSC-selective agent Salinomycin and the Pin1 inhibitor Juglone. Restored expression of the cyclin-dependent kinase inhibitor p16INK4a abrogated the CSC properties of iCSCL-10A cells, by inducing cellular senescence. This study provides some insight into the potential oncogenicity that may arise via cellular reprogramming, and could represent a valuable in vitro model for studying the phenotypic traits of CSCs per se.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755–768. - PubMed
-
- Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. - PubMed
-
- Clevers H. The cancer stem cell: premises, promises and challenges. Nat Med. 2011;17:313–319. - PubMed
-
- Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005;5:275–284. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

