Next-generation sequencing of endoscopic biopsies identifies ARID1A as a tumor-suppressor gene in Barrett's esophagus
- PMID: 23318448
- PMCID: PMC3805724
- DOI: 10.1038/onc.2012.586
Next-generation sequencing of endoscopic biopsies identifies ARID1A as a tumor-suppressor gene in Barrett's esophagus
Abstract
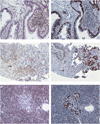
The incidence of Barrett's esophagus (BE)-associated esophageal adenocarcinoma (EAC) is increasing. Next-generation sequencing (NGS) provides an unprecedented opportunity to uncover genomic alterations during BE pathogenesis and progression to EAC, but treatment-naive surgical specimens are scarce. The objective of this study was to establish the feasibility of using widely available endoscopic mucosal biopsies for successful NGS, using samples obtained from a BE 'progressor'. Paired-end whole-genome NGS was performed on the Illumina platform using libraries generated from mucosal biopsies of normal squamous epithelium (NSE), BE and EAC obtained from a patient who progressed to adenocarcinoma during endoscopic surveillance. Selective validation studies, including Sanger sequencing, immunohistochemistry and functional assays, were performed to confirm the NGS findings. NGS identified somatic nonsense mutations of AT-rich interactive domain 1A (SWI like) (ARID1A) and PPIE and an additional 37 missense mutations in BE and/or EAC, which were confirmed by Sanger sequencing. ARID1A mutations were detected in 15% (3/20) high-grade dysplasia (HGD)/EAC patients. Immunohistochemistry performed on an independent archival cohort demonstrated ARID1A protein loss in 0% (0/76), 4.9% (2/40), 14.3% (4/28), 16.0% (8/50) and 12.2% (12/98) of NSE, BE, low-grade dysplasia, HGD and EAC tissues, respectively, and was inversely associated with nuclear p53 accumulation (P=0.028). Enhanced cell growth, proliferation and invasion were observed on ARID1A knockdown in EAC cells. In addition, genes downstream of ARID1A that potentially contribute to the ARID1A knockdown phenotype were identified. Our studies establish the feasibility of using mucosal biopsies for NGS, which should enable the comparative analysis of larger 'progressor' versus 'non-progressor' cohorts. Further, we identify ARID1A as a novel tumor-suppressor gene in BE pathogenesis, reiterating the importance of aberrant chromatin in the metaplasia-dysplasia sequence.
Conflict of interest statement
WRM has participated in Illumina sponsored meetings over the past 4 years and received travel reimbursement and honoraria for presenting at these events. The remaining authors declare no conflict of interest.
Figures



References
-
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. - PubMed
-
- Gilbert EW, Luna RA, Harrison VL, Hunter JG. Barrett’s esophagus: a review of the literature. J Gastrointestinal Surg. 2011;15:708–718. - PubMed
-
- Hvid-Jensen F, Pedersen L, Drewes AM, Sorensen HT, Funch-Jensen P. Incidence of adenocarcinoma among patients with Barrett’s esophagus. N Engl J Med. 2011;365:1375–1383. - PubMed
-
- Yousef F, Cardwell C, Cantwell MM, Galway K, Johnston BT, Murray L. The incidence of esophageal cancer and high-grade dysplasia in Barrett’s esophagus: a systematic review and meta-analysis. Am J Epidemiol. 2008;168:237–249. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

