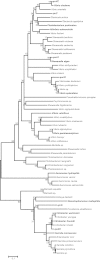
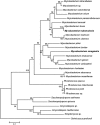
Phylogenetic analysis of chromosomally determined qnr and related proteins
- PMID: 23318805
- PMCID: PMC3623327
- DOI: 10.1128/AAC.02080-12
Phylogenetic analysis of chromosomally determined qnr and related proteins
Abstract
qnr genes were discovered on plasmids by their ability to reduce quinolone susceptibility, but homologs can be found in the genomes of at least 92 Gram-negative, Gram-positive, and strictly anaerobic bacterial species. The related pentapeptide repeat protein-encoding mfpA gene is present in the genome of at least 19 species of Mycobacterium and 10 other Actinobacteria species. The native function of these genes is not yet known.
Figures

References
-
- Martínez-Martínez L, Pascual A, Jacoby GA. 1998. Quinolone resistance from a transferable plasmid. Lancet 351:797–799 - PubMed
-
- Xiong X, Bromley EH, Oelschlaeger P, Woolfson DN, Spencer J. 2011. Structural insights into quinolone antibiotic resistance mediated by pentapeptide repeat proteins: conserved surface loops direct the activity of a Qnr protein from a gram-negative bacterium. Nucleic Acids Res. 39:3917–3927 - PMC - PubMed
-
- Sun HI, Jeong DU, Lee JH, Wu X, Park KS, Lee JJ, Jeong BC, Lee SH. 2010. A novel family (QnrAS) of plasmid-mediated quinolone resistance determinant. Int. J. Antimicrob. Agents 36:578–579 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

