Mutations in interaction surfaces differentially impact E. coli Hfq association with small RNAs and their mRNA targets
- PMID: 23318956
- PMCID: PMC3640674
- DOI: 10.1016/j.jmb.2013.01.006
Mutations in interaction surfaces differentially impact E. coli Hfq association with small RNAs and their mRNA targets
Abstract
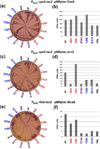
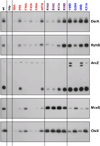
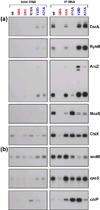
The RNA chaperone protein Hfq is required for the function of all small RNAs (sRNAs) that regulate mRNA stability or translation by limited base pairing in Escherichia coli. While there have been numerous in vitro studies to characterize Hfq activity and the importance of specific residues, there has been only limited characterization of Hfq mutants in vivo. Here, we use a set of reporters as well as co-immunoprecipitation to examine 14 Hfq mutants expressed from the E. coli chromosome. The majority of the proximal face residues, as expected, were important for the function of sRNAs. The failure of sRNAs to regulate target mRNAs in these mutants can be explained by reduced sRNA accumulation. Two of the proximal mutants, D9A and F39A, acted differently from the others in that they had mixed effects on different sRNA/mRNA pairs and, in the case of F39A, showed differential sRNA accumulation. Mutations of charged residues at the rim of Hfq interfered with positive regulation and gave mixed effects for negative regulation. Some, but not all, sRNAs accumulated to lower levels in rim mutants, suggesting qualitative differences in how individual sRNAs are affected by Hfq. The distal face mutants were expected to disrupt binding of ARN motifs found in mRNAs. They were more defective for positive regulation than negative regulation at low mRNA expression, but the defects could be suppressed by higher levels of mRNA expression. We discuss the implications of these observations for Hfq binding to RNA and mechanisms of action.
Keywords: ArcZ; BSA; ChiX; DsrA; LB; Luria–Bertani; McaS; NSWB; RyhB; bovine serum albumin; co-IP; co-immunoprecipitation; non-stringent wash buffer; sRNA; small RNA; wild type; wt.
Published by Elsevier Ltd.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

