Dispersed sites of HIV Vif-dependent polyubiquitination in the DNA deaminase APOBEC3F
- PMID: 23318957
- PMCID: PMC3602375
- DOI: 10.1016/j.jmb.2013.01.010
Dispersed sites of HIV Vif-dependent polyubiquitination in the DNA deaminase APOBEC3F
Abstract
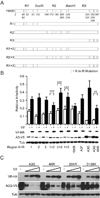
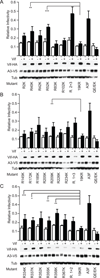
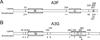
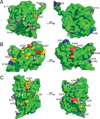
APOBEC3F (A3F) and APOBEC3G (A3G) are DNA cytosine deaminases that potently restrict human immunodeficiency virus type 1 replication when the virus is deprived of its accessory protein Vif (virion infectivity factor). Vif counteracts these restriction factors by recruiting A3F and A3G to an E3 ubiquitin (Ub) ligase complex that mediates their polyubiquitination (polyUb) and proteasomal degradation. While previous efforts have identified single amino acid residues in APOBEC3 proteins required for Vif recognition, less is known about the downstream Ub acceptor sites that are targeted. One prior report identified a cluster of polyubiquitinated residues in A3G and proposed an antiparallel model of A3G interaction with the Vif-E3 Ub ligase complex wherein Vif binding at one terminus of A3G orients the opposite terminus for polyUb [Iwatani et al. (2009). Proc. Natl. Acad. Sci. USA, 106, 19539-19544]. To test the generalizability of this model, we carried out a complete mutagenesis of the lysine residues in A3F and used a complementary, unbiased proteomic approach to identify Ub acceptor sites targeted by Vif. Our data indicate that internal lysines are the dominant Ub acceptor sites in both A3F and A3G. In contrast with the proposed antiparallel model, however, we find that the Vif-dependent polyUb of A3F and A3G can occur at multiple acceptor sites dispersed along predicted lysine-enriched surfaces of both the N- and C-terminal deaminase domains. These data suggest an alternative model for binding of APOBEC3 proteins to the Vif-E3 Ub ligase complex and diminish enthusiasm for the amenability of APOBEC3 Ub acceptor sites to therapeutic intervention.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Figures

References
-
- Chiu YL, Greene WC. The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu Rev Immunol. 2008;26:317–353. - PubMed
-
- Jäger S, Kim DY, Hultquist JF, Shindo K, Larue RS, Kwon E, Li M, Anderson BD, Yen L, Stanley D, Mahon C, Kane J, Franks-Skiba K, Cimermancic P, Burlingame A, Sali A, Craik CS, Harris RS, Gross JD, Krogan NJ. Vif hijacks CBFβ to degrade APOBEC3G and promote HIV-1 infection. Nature. 2011;481:371–375. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

