Hypoxia-inducible factor-dependent breast cancer-mesenchymal stem cell bidirectional signaling promotes metastasis
- PMID: 23318994
- PMCID: PMC3533298
- DOI: 10.1172/JCI64993
Hypoxia-inducible factor-dependent breast cancer-mesenchymal stem cell bidirectional signaling promotes metastasis
Erratum in
- J Clin Invest. 2013 Mar 1;123(3):1402
Abstract
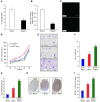
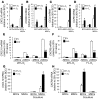
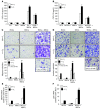
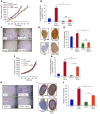
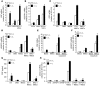
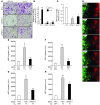
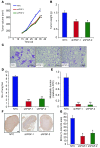
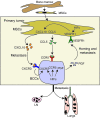
Metastasis involves critical interactions between cancer and stromal cells. Intratumoral hypoxia promotes metastasis through activation of hypoxia-inducible factors (HIFs). We demonstrate that HIFs mediate paracrine signaling between breast cancer cells (BCCs) and mesenchymal stem cells (MSCs) to promote metastasis. In a mouse orthotopic implantation model, MSCs were recruited to primary breast tumors and promoted BCC metastasis to LNs and lungs in a HIF-dependent manner. Coculture of MSCs with BCCs augmented HIF activity in BCCs. Additionally, coculture induced expression of the chemokine CXCL10 in MSCs and the cognate receptor CXCR3 in BCCs, which was augmented by hypoxia. CXCR3 expression was blocked in cocultures treated with neutralizing antibody against CXCL10. Conversely, CXCL10 expression was blocked in MSCs cocultured with BCCs that did not express CXCR3 or HIFs. MSC coculture did not enhance the metastasis of HIF-deficient BCCs. BCCs and MSCs expressed placental growth factor (PGF) and its cognate receptor VEGFR1, respectively, in a HIF-dependent manner, and CXCL10 expression by MSCs was dependent on PGF expression by BCCs. PGF promoted metastasis of BCCs and also facilitated homing of MSCs to tumors. Thus, HIFs mediate complex and bidirectional paracrine signaling between BCCs and MSCs that stimulates breast cancer metastasis.
Figures