Association testing of the mitochondrial genome using pedigree data
- PMID: 23319385
- PMCID: PMC4171957
- DOI: 10.1002/gepi.21706
Association testing of the mitochondrial genome using pedigree data
Abstract
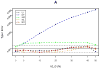
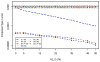
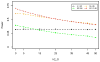
In humans, mitochondria contain their own DNA (mtDNA) that is inherited exclusively from the mother. The mitochondrial genome encodes 13 polypeptides that are components of oxidative phosphorylation to produce energy. Any disruption in these genes might interfere with energy production and thus contribute to metabolic derangement. Mitochondria also regulate several important cellular activities including cell death and calcium homeostasis. Aided by sharply declining costs of high-density genotyping, hundreds of mitochondrial variants will soon be available in several cohorts with pedigree structures. Association testing of mitochondrial variants with disease traits using pedigree data raises unique challenges because of the difficulty in separating the effects of nuclear and mitochondrial genomes, which display different modes of inheritance. Failing to correctly account for these effects might decrease power or inflate type I error in association tests. In this report, we sought to identify the best strategy for association testing of mitochondrial variants when genotype and phenotype data are available in pedigrees. We proposed several strategies to account for polygenic effects of the nuclear and mitochondrial genomes and we performed extensive simulation studies to evaluate type I error and power of these strategies. In addition, we proposed two permutation tests to obtain empirical P values for these strategies. Furthermore, we applied two of the analytical strategies to association analysis of 196 mitochondrial variants with blood pressure and fasting blood glucose in the pedigree rich, Framingham Heart Study. Finally, we discussed strategies for study design, genotyping, and data cleaning in association testing of mtDNA in pedigrees.
© 2013 Wiley Periodicals, Inc.
Conflict of interest statement
The authors declare that there is no conflict of interest.
Figures

Similar articles
-
Pedigree models for complex human traits involving the mitochondrial genome.Am J Hum Genet. 1993 Dec;53(6):1320-37. Am J Hum Genet. 1993. PMID: 8250048 Free PMC article.
-
Family-based association tests for genomewide association scans.Am J Hum Genet. 2007 Nov;81(5):913-26. doi: 10.1086/521580. Epub 2007 Sep 18. Am J Hum Genet. 2007. PMID: 17924335 Free PMC article.
-
Quantitative pedigree analysis and mitochondrial DNA sequence variants in adults with cyclic vomiting syndrome.BMC Gastroenterol. 2014 Oct 21;14:181. doi: 10.1186/1471-230X-14-181. BMC Gastroenterol. 2014. PMID: 25332060 Free PMC article.
-
Mitochondrial DNA, nuclear context, and the risk for carcinogenesis.Environ Mol Mutagen. 2019 Jun;60(5):455-462. doi: 10.1002/em.22169. Epub 2018 Jan 14. Environ Mol Mutagen. 2019. PMID: 29332303 Free PMC article. Review.
-
Mitochondrial DNA genetics and the heteroplasmy conundrum in evolution and disease.Cold Spring Harb Perspect Biol. 2013 Nov 1;5(11):a021220. doi: 10.1101/cshperspect.a021220. Cold Spring Harb Perspect Biol. 2013. PMID: 24186072 Free PMC article. Review.
Cited by
-
Association of mitochondrial DNA copy number with cardiometabolic diseases.Cell Genom. 2021 Oct 13;1(1):100006. doi: 10.1016/j.xgen.2021.100006. Cell Genom. 2021. PMID: 35036986 Free PMC article.
-
Relations of mitochondrial genetic variants to measures of vascular function.Mitochondrion. 2018 May;40:51-57. doi: 10.1016/j.mito.2017.10.001. Epub 2017 Oct 7. Mitochondrion. 2018. PMID: 28993255 Free PMC article.
-
Revisiting heritability accounting for shared environmental effects and maternal inheritance.Hum Genet. 2015 Feb;134(2):169-79. doi: 10.1007/s00439-014-1505-6. Epub 2014 Nov 9. Hum Genet. 2015. PMID: 25381465 Free PMC article. Clinical Trial.
-
Associations of Mitochondrial and Nuclear Mitochondrial Variants and Genes with Seven Metabolic Traits.Am J Hum Genet. 2019 Jan 3;104(1):112-138. doi: 10.1016/j.ajhg.2018.12.001. Epub 2018 Dec 27. Am J Hum Genet. 2019. PMID: 30595373 Free PMC article.
-
MaGelLAn 1.0: a software to facilitate quantitative and population genetic analysis of maternal inheritance by combination of molecular and pedigree information.Genet Sel Evol. 2016 Sep 10;48(1):65. doi: 10.1186/s12711-016-0242-9. Genet Sel Evol. 2016. PMID: 27613390 Free PMC article.
References
-
- Benn M, Schwartz M, Nordestgaard BG, Tybjaerg-Hansen A. Mitochondrial haplogroups: ischemic cardiovascular disease, other diseases, mortality, and longevity in the general population. Circulation. 2008;117(19):2492–501. - PubMed
-
- Byrne EM, McRae AF, Duffy DL, Zhao ZZ, Martin NG, Whitfield JB, Visscher PM, Montgomery GW. Family-based mitochondrial association study of traits related to type 2 diabetes and the metabolic syndrome in adolescents. Diabetologia. 2009;52(11):2359–68. - PubMed
-
- Dawber TR, Kannel WB, Lyell LP. An approach to longitudinal studies in a community: the Framingham Study. Ann N Y Acad Sci. 1963;107:539–56. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

