Analysis of Ca2+ signaling motifs that regulate proton signaling through the Na+/H+ exchanger NHX-7 during a rhythmic behavior in Caenorhabditis elegans
- PMID: 23319594
- PMCID: PMC3581405
- DOI: 10.1074/jbc.M112.434852
Analysis of Ca2+ signaling motifs that regulate proton signaling through the Na+/H+ exchanger NHX-7 during a rhythmic behavior in Caenorhabditis elegans
Abstract
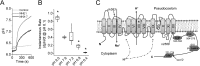
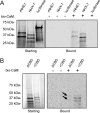
Membrane proton transporters contribute to pH homeostasis but have also been shown to transmit information between cells in close proximity through regulated proton secretion. For example, the nematode intestinal Na(+)/H(+) exchanger NHX-7 causes adjacent muscle cells to contract by transiently acidifying the extracellular space between the intestine and muscle. NHX-7 operates during a Ca(2+)-dependent rhythmic behavior and contains several conserved motifs for regulation by Ca(2+) input, including motifs for calmodulin and phosphatidylinositol 4,5-bisphosphate binding, protein kinase C- and calmodulin-dependent protein kinase type II phosphorylation, and a binding site for calcineurin homologous protein. Here, we tested the idea that Ca(2+) input differentiates proton signaling from pH housekeeping activity. Each of these motifs was mutated, and their contribution to NHX-7 function was assessed. These functions included pH recovery from acidification in cells in culture expressing recombinant NHX-7, extracellular acidification measured during behavior in live moving worms, and muscle contraction strength as a result of this acidification. Our data suggest that multiple levels of Ca(2+) input regulate NHX-7, whose transport capacity normally exceeds the minimum necessary to cause muscle contraction. Furthermore, extracellular acidification limits NHX-7 proton transport through feedback inhibition, likely to prevent metabolic acidosis from occurring. Our findings are consistent with an integrated network whereby both Ca(2+) and pH contribute to proton signaling. Finally, our results obtained by expressing rat NHE1 in Caenorhabditis elegans suggest that a conserved mechanism of regulation may contribute to cell-cell communication or proton signaling by Na(+)/H(+) exchangers in mammals.
Figures

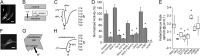
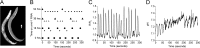
Similar articles
-
A reduction in intestinal cell pHi due to loss of the Caenorhabditis elegans Na+/H+ exchanger NHX-2 increases life span.J Biol Chem. 2003 Nov 7;278(45):44657-66. doi: 10.1074/jbc.M307351200. Epub 2003 Aug 25. J Biol Chem. 2003. PMID: 12939266
-
Calcineurin homologous proteins regulate the membrane localization and activity of sodium/proton exchangers in C. elegans.Am J Physiol Cell Physiol. 2016 Feb 1;310(3):C233-42. doi: 10.1152/ajpcell.00291.2015. Epub 2015 Nov 11. Am J Physiol Cell Physiol. 2016. PMID: 26561640
-
Oscillatory transepithelial H(+) flux regulates a rhythmic behavior in C. elegans.Curr Biol. 2008 Feb 26;18(4):297-302. doi: 10.1016/j.cub.2008.01.054. Curr Biol. 2008. PMID: 18291648 Free PMC article.
-
Regulation of the cardiac Na⁺/H⁺ exchanger in health and disease.J Mol Cell Cardiol. 2013 Aug;61:68-76. doi: 10.1016/j.yjmcc.2013.02.007. Epub 2013 Feb 18. J Mol Cell Cardiol. 2013. PMID: 23429007 Review.
-
Structure and function of the NHE1 isoform of the Na+/H+ exchanger.Biochem Cell Biol. 2002;80(5):499-508. doi: 10.1139/o02-151. Biochem Cell Biol. 2002. PMID: 12440691 Review.
Cited by
-
Human calmodulin mutations cause arrhythmia and affect neuronal function in C. elegans.Hum Mol Genet. 2023 Jun 5;32(12):2068-2083. doi: 10.1093/hmg/ddad042. Hum Mol Genet. 2023. PMID: 36920509 Free PMC article.
-
Distinct roles for two Caenorhabditis elegans acid-sensing ion channels in an ultradian clock.Elife. 2022 Jun 6;11:e75837. doi: 10.7554/eLife.75837. Elife. 2022. PMID: 35666106 Free PMC article.
-
Transcriptomic analysis unravels the molecular response of Lonicera japonica leaves to chilling stress.Front Plant Sci. 2022 Dec 22;13:1092857. doi: 10.3389/fpls.2022.1092857. eCollection 2022. Front Plant Sci. 2022. PMID: 36618608 Free PMC article.
References
-
- Casey J. R., Grinstein S., Orlowski J. (2010) Sensors and regulators of intracellular pH. Nat. Rev. Mol. Cell Biol. 11, 50–61 - PubMed
-
- Alper S. L. (2002) Genetic diseases of acid-base transporters. Annu. Rev. Physiol. 64, 899–923 - PubMed
-
- Martínez-Zaguilán R., Seftor E. A., Seftor R. E., Chu Y. W., Gillies R. J., Hendrix M. J. (1996) Acidic pH enhances the invasive behavior of human melanoma cells. Clin. Exp. Metastasis 14, 176–186 - PubMed
-
- Ludwig M. G., Vanek M., Guerini D., Gasser J. A., Jones C. E., Junker U., Hofstetter H., Wolf R. M., Seuwen K. (2003) Proton-sensing G-protein-coupled receptors. Nature 425, 93–98 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

