Structural basis for cytosolic double-stranded RNA surveillance by human oligoadenylate synthetase 1
- PMID: 23319625
- PMCID: PMC3562804
- DOI: 10.1073/pnas.1218528110
Structural basis for cytosolic double-stranded RNA surveillance by human oligoadenylate synthetase 1
Abstract
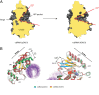
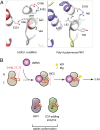
The human sensor of double-stranded RNA (dsRNA) oligoadenylate synthetase 1 (hOAS1) polymerizes ATP into 2',5'-linked iso-RNA (2-5A) involved in innate immunity, cell cycle, and differentiation. We report the crystal structure of hOAS1 in complex with dsRNA and 2'-deoxy ATP at 2.7 Å resolution, which reveals the mechanism of cytoplasmic dsRNA recognition and activation of oligoadenylate synthetases. Human OAS1 recognizes dsRNA using a previously uncharacterized protein/RNA interface that forms via a conformational change induced by binding of dsRNA. The protein/RNA interface involves two minor grooves and has no sequence-specific contacts, with the exception of a single hydrogen bond between the -NH(2) group of nucleobase G17 and the carbonyl oxygen of serine 56. Using a biochemical readout, we show that hOAS1 undergoes more than 20,000-fold activation upon dsRNA binding and that canonical or GU-wobble substitutions produce dsRNA mutants that retain either full or partial activity, in agreement with the crystal structure. Ultimately, the binding of dsRNA promotes an elaborate conformational rearrangement in the N-terminal lobe of hOAS1, which brings residues D75, D77, and D148 into proximity and creates coordination geometry for binding of two catalytic Mg(2+) ions and ATP. The assembly of this critical active-site structure provides the gate that couples binding of dsRNA to the production and downstream functions of 2-5A.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Kodym R, Kodym E, Story MD. 2′-5′-Oligoadenylate synthetase is activated by a specific RNA sequence motif. Biochem Biophys Res Commun. 2009;388(2):317–322. - PubMed
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

