Explaining why Gleevec is a specific and potent inhibitor of Abl kinase
- PMID: 23319661
- PMCID: PMC3562763
- DOI: 10.1073/pnas.1214330110
Explaining why Gleevec is a specific and potent inhibitor of Abl kinase
Abstract
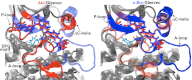
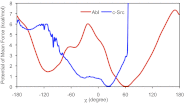
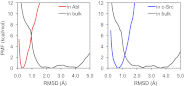
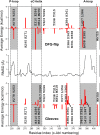
Tyrosine kinases present attractive drug targets for specific types of cancers. Gleevec, a well-known therapeutic agent against chronic myelogenous leukemia, is an effective inhibitor of Abl tyrosine kinase. However, Gleevec fails to inhibit closely homologous tyrosine kinases, such as c-Src. Because many structural features of the binding site are conserved, the molecular determinants responsible for binding specificity are not immediately apparent. Some have attributed the difference in binding specificity of Gleevec to subtle variations in ligand-protein interactions (binding affinity control), whereas others have proposed that it is the conformation of the DFG motif, in which ligand binding is only accessible to Abl and not to c-Src (conformational selection control). To address this issue, the absolute binding free energy was computed using all-atom molecular dynamics simulations with explicit solvent. The results of the free energy simulations are in good agreement with experiments, thereby enabling a meaningful decomposition of the binding free energy to elucidate the factors controlling Gleevec's binding specificity. The latter is shown to be controlled by a conformational selection mechanism and also by differences in key van der Waals interactions responsible for the stabilization of Gleevec in the binding pocket of Abl.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Computational analysis of the binding specificity of Gleevec to Abl, c-Kit, Lck, and c-Src tyrosine kinases.J Am Chem Soc. 2013 Oct 2;135(39):14741-53. doi: 10.1021/ja405939x. Epub 2013 Sep 20. J Am Chem Soc. 2013. PMID: 24001034 Free PMC article.
-
Computational study of Gleevec and G6G reveals molecular determinants of kinase inhibitor selectivity.J Am Chem Soc. 2014 Oct 22;136(42):14753-62. doi: 10.1021/ja504146x. Epub 2014 Oct 7. J Am Chem Soc. 2014. PMID: 25243930 Free PMC article.
-
Energetic dissection of Gleevec's selectivity toward human tyrosine kinases.Nat Struct Mol Biol. 2014 Oct;21(10):848-53. doi: 10.1038/nsmb.2891. Epub 2014 Sep 14. Nat Struct Mol Biol. 2014. PMID: 25218445 Free PMC article.
-
New Bcr-Abl inhibitors in chronic myeloid leukemia: keeping resistance in check.Expert Opin Investig Drugs. 2008 Jun;17(6):865-78. doi: 10.1517/13543784.17.6.865. Expert Opin Investig Drugs. 2008. PMID: 18491988 Review.
-
Mechanisms of resistance to imatinib mesylate in Bcr-Abl-positive leukemias.Curr Opin Oncol. 2002 Nov;14(6):616-20. doi: 10.1097/00001622-200211000-00005. Curr Opin Oncol. 2002. PMID: 12409651 Review.
Cited by
-
Kinase dynamics. Using ancient protein kinases to unravel a modern cancer drug's mechanism.Science. 2015 Feb 20;347(6224):882-6. doi: 10.1126/science.aaa1823. Science. 2015. PMID: 25700521 Free PMC article.
-
Integrative X-ray Structure and Molecular Modeling for the Rationalization of Procaspase-8 Inhibitor Potency and Selectivity.ACS Chem Biol. 2020 Feb 21;15(2):575-586. doi: 10.1021/acschembio.0c00019. Epub 2020 Jan 23. ACS Chem Biol. 2020. PMID: 31927936 Free PMC article.
-
Molecular basis for differential recognition of an allosteric inhibitor by receptor tyrosine kinases.Proteins. 2024 Aug;92(8):905-922. doi: 10.1002/prot.26685. Epub 2024 Mar 20. Proteins. 2024. PMID: 38506327 Free PMC article.
-
An activation-based high throughput screen identifies caspase-10 inhibitors.RSC Chem Biol. 2025 Feb 4;6(4):604-617. doi: 10.1039/d5cb00017c. eCollection 2025 Apr 2. RSC Chem Biol. 2025. PMID: 40013156 Free PMC article.
-
Structural propensities of kinase family proteins from a Potts model of residue co-variation.Protein Sci. 2016 Aug;25(8):1378-84. doi: 10.1002/pro.2954. Epub 2016 Jun 26. Protein Sci. 2016. PMID: 27241634 Free PMC article.
References
-
- Hosfield DJ, Mol CD. Targeting inactive kinases: Structure as a foundation for cancer drug discovery. In: Neidle S, editor. Cancer Drug Design and Discovery. San Diego: Elsevier Science & Technology Books; 2008. pp. 229–252.
-
- Capdeville R, Buchdunger E, Zimmermann J, Matter A. Glivec (STI571, imatinib), a rationally developed, targeted anticancer drug. Nat Rev Drug Discov. 2002;1(7):493–502. - PubMed
-
- Nowell PC, Hungerford DA. Chromosome studies on normal and leukemic human leukocytes. J Natl Cancer Inst. 1960;25:85–109. - PubMed
-
- Rowley JD. Letter: A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243(5405):290–293. - PubMed
-
- Schindler T, et al. Structural mechanism for STI-571 inhibition of abelson tyrosine kinase. Science. 2000;289(5486):1938–1942. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

