FOXP3 regulates sensitivity of cancer cells to irradiation by transcriptional repression of BRCA1
- PMID: 23319807
- PMCID: PMC3815443
- DOI: 10.1158/0008-5472.CAN-12-2481
FOXP3 regulates sensitivity of cancer cells to irradiation by transcriptional repression of BRCA1
Abstract
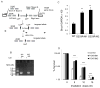
FOXP3 is an X-linked tumor suppressor gene and a master regulator in T regulatory cell function. This gene has been found to be mutated frequently in breast and prostate cancers and to inhibit tumor cell growth, but its functional significance in DNA repair has not been studied. We found that FOXP3 silencing stimulates homologous recombination-mediated DNA repair and also repair of γ-irradiation-induced DNA damage. Expression profiling and chromatin-immunoprecipitation analyses revealed that FOXP3 regulated the BRCA1-mediated DNA repair program. Among 48 FOXP3-regulated DNA repair genes, BRCA1 and 12 others were direct targets of FOXP3 transcriptional control. Site-specific interaction of FOXP3 with the BRCA1 promoter repressed its transcription. Somatic FOXP3 mutants identified in breast cancer samples had reduced BRCA1 repressor activity, whereas FOXP3 silencing and knock-in of a prostate cancer-derived somatic FOXP3 mutant increased the radioresistance of cancer cells. Together our findings provide a missing link between FOXP3 function and DNA repair programs.
©2012 AACR.
Conflict of interest statement
The authors have no conflict of interest in this study.
Figures





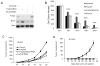
Similar articles
-
Hypoxia-induced down-regulation of BRCA1 expression by E2Fs.Cancer Res. 2005 Dec 15;65(24):11597-604. doi: 10.1158/0008-5472.CAN-05-2119. Cancer Res. 2005. PMID: 16357170
-
Downregulation of Ezh2 methyltransferase by FOXP3: new insight of FOXP3 into chromatin remodeling?Biochim Biophys Acta. 2013 Oct;1833(10):2190-200. doi: 10.1016/j.bbamcr.2013.05.014. Epub 2013 May 17. Biochim Biophys Acta. 2013. PMID: 23688634
-
BRCA1 represses amphiregulin gene expression.Cancer Res. 2010 Feb 1;70(3):996-1005. doi: 10.1158/0008-5472.CAN-09-2842. Epub 2010 Jan 26. Cancer Res. 2010. PMID: 20103632 Free PMC article.
-
Therapeutic exploitation of tumor cell defects in homologous recombination.Anticancer Agents Med Chem. 2008 May;8(4):448-60. doi: 10.2174/187152008784220267. Anticancer Agents Med Chem. 2008. PMID: 18473729 Review.
-
BRCA1 role in the mitigation of radiotoxicity and chromosomal instability through repair of clustered DNA lesions.Chem Biol Interact. 2010 Nov 5;188(2):350-8. doi: 10.1016/j.cbi.2010.03.046. Epub 2010 Apr 4. Chem Biol Interact. 2010. PMID: 20371364 Review.
Cited by
-
FOXP3 over-expression inhibits melanoma tumorigenesis via effects on proliferation and apoptosis.Oncotarget. 2014 Jan 15;5(1):264-76. doi: 10.18632/oncotarget.1600. Oncotarget. 2014. PMID: 24406338 Free PMC article.
-
FOXP3 promotes tumor growth and metastasis by activating Wnt/β-catenin signaling pathway and EMT in non-small cell lung cancer.Mol Cancer. 2017 Jul 17;16(1):124. doi: 10.1186/s12943-017-0700-1. Mol Cancer. 2017. PMID: 28716029 Free PMC article.
-
Forkhead Box Protein P3 (FOXP3) Represses ATF3 Transcriptional Activity.Int J Mol Sci. 2021 Oct 22;22(21):11400. doi: 10.3390/ijms222111400. Int J Mol Sci. 2021. PMID: 34768829 Free PMC article.
-
Foxp3 expression in T regulatory cells and other cell lineages.Cancer Immunol Immunother. 2014 Sep;63(9):869-76. doi: 10.1007/s00262-014-1581-4. Epub 2014 Jul 26. Cancer Immunol Immunother. 2014. PMID: 25063364 Free PMC article. Review.
-
BRCA1 Mutation: A Predictive Marker for Radiation Therapy?Int J Radiat Oncol Biol Phys. 2015 Oct 1;93(2):281-93. doi: 10.1016/j.ijrobp.2015.05.037. Int J Radiat Oncol Biol Phys. 2015. PMID: 26383678 Free PMC article. Review.
References
-
- Begg AC, Stewart FA, Vens C. Strategies to improve radiotherapy with targeted drugs. Nat Rev Cancer. 2011;11:239–53. - PubMed
-
- Delaney G, Jacob S, Featherstone C, Barton M. The role of radiotherapy in cancer treatment: estimating optimal utilization from a review of evidence-based clinical guidelines. Cancer. 2005;104:1129–37. - PubMed
-
- Baumann M, Krause M, Hill R. Exploring the role of cancer stem cells in radioresistance. Nat Rev Cancer. 2008;8:545–54. - PubMed
-
- Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–60. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

