SON protein regulates GATA-2 through transcriptional control of the microRNA 23a~27a~24-2 cluster
- PMID: 23322776
- PMCID: PMC3581430
- DOI: 10.1074/jbc.M112.447227
SON protein regulates GATA-2 through transcriptional control of the microRNA 23a~27a~24-2 cluster
Abstract
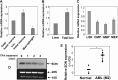
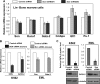
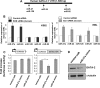
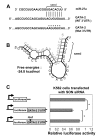
SON is a DNA- and RNA-binding protein localized in nuclear speckles. Although its function in RNA splicing for effective cell cycle progression and genome stability was recently unveiled, other mechanisms of SON functions remain unexplored. Here, we report that SON regulates GATA-2, a key transcription factor involved in hematopoietic stem cell maintenance and differentiation. SON is highly expressed in undifferentiated hematopoietic stem/progenitor cells and leukemic blasts. SON knockdown leads to significant depletion of GATA-2 protein with marginal down-regulation of GATA-2 mRNA. We show that miR-27a is up-regulated upon SON knockdown and targets the 3'-UTR of GATA-2 mRNA in hematopoietic cells. Up-regulation of miR-27a was due to activation of the promoter of the miR-23a∼27a∼24-2 cluster, suggesting that SON suppresses this promoter to lower the microRNAs from this cluster. Our data revealed a previously unidentified role of SON in microRNA production via regulating the transcription process, thereby modulating GATA-2 at the protein level during hematopoietic differentiation.
Figures

References
-
- Berdichevskii F. B., Chumakov I. M., Kiselev L. L. (1988) Decoding the primary structure of the son3 region in human genome: identification of a new protein with unusual structure and homology with DNA-binding proteins. Mol. Biol. 22, 794–801 - PubMed
-
- Chumakov I. M., Berdichevskii F. B., Sokolova N. V., Reznikov M. V., Prasolov V. S. (1991) Identification of a protein product of a novel human gene SON and the biological effect upon administering a changed form of this gene into mammalian cells. Mol. Biol. 25, 731–739 - PubMed
-
- Greenhalf W., Lee J., Chaudhuri B. (1999) A selection system for human apoptosis inhibitors using yeast. Yeast 15, 1307–1321 - PubMed
-
- Sun C. T., Lo W. Y., Wang I. H., Lo Y. H., Shiou S. R., Lai C. K., Ting L. P. (2001) Transcription repression of human hepatitis B virus genes by negative regulatory element-binding protein/SON. J. Biol. Chem. 276, 24059–24067 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

