Micrometric segregation of fluorescent membrane lipids: relevance for endogenous lipids and biogenesis in erythrocytes
- PMID: 23322884
- PMCID: PMC3605983
- DOI: 10.1194/jlr.M034314
Micrometric segregation of fluorescent membrane lipids: relevance for endogenous lipids and biogenesis in erythrocytes
Erratum in
- J Lipid Res. 2013 Jul;54(7):2029
Abstract
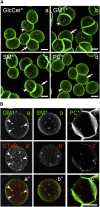
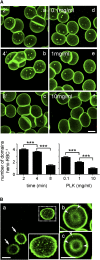
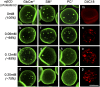
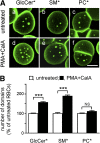
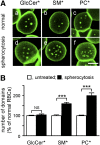
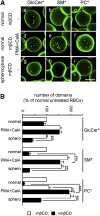
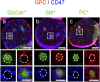
Micrometric membrane lipid segregation is controversial. We addressed this issue in attached erythrocytes and found that fluorescent boron dipyrromethene (BODIPY) analogs of glycosphingolipids (GSLs) [glucosylceramide (BODIPY-GlcCer) and monosialotetrahexosylganglioside (GM1BODIPY)], sphingomyelin (BODIPY-SM), and phosphatidylcholine (BODIPY-PC inserted into the plasma membrane spontaneously gathered into distinct submicrometric domains. GM1BODIPY domains colocalized with endogenous GM1 labeled by cholera toxin. All BODIPY-lipid domains disappeared upon erythrocyte stretching, indicating control by membrane tension. Minor cholesterol depletion suppressed BODIPY-SM and BODIPY-PC but preserved BODIPY-GlcCer domains. Each type of domain exchanged constituents but assumed fixed positions, suggesting self-clustering and anchorage to spectrin. Domains showed differential association with 4.1R versus ankyrin complexes upon antibody patching. BODIPY-lipid domains also responded differentially to uncoupling at 4.1R complexes [protein kinase C (PKC) activation] and ankyrin complexes (in spherocytosis, a membrane fragility disease). These data point to micrometric compartmentation of polar BODIPY-lipids modulated by membrane tension, cholesterol, and differential association to the two nonredundant membrane:spectrin anchorage complexes. Micrometric compartmentation might play a role in erythrocyte membrane deformability and fragility.
Figures

References
-
- Singer S. J., Nicolson G. L. 1972. The fluid mosaic model of the structure of cell membranes. Science. 175: 720–731. - PubMed
-
- Lingwood D., Simons K. 2010. Lipid rafts as a membrane-organizing principle. Science. 327: 46–50. - PubMed
-
- Pike L. J. 2006. Rafts defined: a report on the Keystone Symposium on Lipid Rafts and Cell Function. J. Lipid Res. 47: 1597–1598. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

