A detailed error analysis of 13 kernel methods for protein-protein interaction extraction
- PMID: 23323857
- PMCID: PMC3680070
- DOI: 10.1186/1471-2105-14-12
A detailed error analysis of 13 kernel methods for protein-protein interaction extraction
Abstract
Background: Kernel-based classification is the current state-of-the-art for extracting pairs of interacting proteins (PPIs) from free text. Various proposals have been put forward, which diverge especially in the specific kernel function, the type of input representation, and the feature sets. These proposals are regularly compared to each other regarding their overall performance on different gold standard corpora, but little is known about their respective performance on the instance level.
Results: We report on a detailed analysis of the shared characteristics and the differences between 13 current methods using five PPI corpora. We identified a large number of rather difficult (misclassified by most methods) and easy (correctly classified by most methods) PPIs. We show that kernels using the same input representation perform similarly on these pairs and that building ensembles using dissimilar kernels leads to significant performance gain. However, our analysis also reveals that characteristics shared between difficult pairs are few, which lowers the hope that new methods, if built along the same line as current ones, will deliver breakthroughs in extraction performance.
Conclusions: Our experiments show that current methods do not seem to do very well in capturing the shared characteristics of positive PPI pairs, which must also be attributed to the heterogeneity of the (still very few) available corpora. Our analysis suggests that performance improvements shall be sought after rather in novel feature sets than in novel kernel functions.
Figures
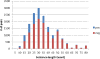
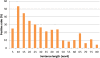
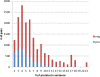
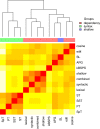
Similar articles
-
A comprehensive benchmark of kernel methods to extract protein-protein interactions from literature.PLoS Comput Biol. 2010 Jul 1;6(7):e1000837. doi: 10.1371/journal.pcbi.1000837. PLoS Comput Biol. 2010. PMID: 20617200 Free PMC article.
-
Neighborhood hash graph kernel for protein-protein interaction extraction.J Biomed Inform. 2011 Dec;44(6):1086-92. doi: 10.1016/j.jbi.2011.08.011. Epub 2011 Aug 23. J Biomed Inform. 2011. PMID: 21884822
-
Distributed smoothed tree kernel for protein-protein interaction extraction from the biomedical literature.PLoS One. 2017 Nov 3;12(11):e0187379. doi: 10.1371/journal.pone.0187379. eCollection 2017. PLoS One. 2017. PMID: 29099838 Free PMC article.
-
Targeting Virus-host Protein Interactions: Feature Extraction and Machine Learning Approaches.Curr Drug Metab. 2019;20(3):177-184. doi: 10.2174/1389200219666180829121038. Curr Drug Metab. 2019. PMID: 30156155 Review.
-
Techniques for the Analysis of Protein-Protein Interactions in Vivo.Plant Physiol. 2016 Jun;171(2):727-58. doi: 10.1104/pp.16.00470. Epub 2016 Apr 25. Plant Physiol. 2016. PMID: 27208310 Free PMC article. Review.
Cited by
-
Extracting drug-enzyme relation from literature as evidence for drug drug interaction.J Biomed Semantics. 2016 Mar 7;7:11. doi: 10.1186/s13326-016-0052-6. eCollection 2016. J Biomed Semantics. 2016. PMID: 26955465 Free PMC article.
-
PEDL: extracting protein-protein associations using deep language models and distant supervision.Bioinformatics. 2020 Jul 1;36(Suppl_1):i490-i498. doi: 10.1093/bioinformatics/btaa430. Bioinformatics. 2020. PMID: 32657389 Free PMC article.
-
Translating cancer genomics into precision medicine with artificial intelligence: applications, challenges and future perspectives.Hum Genet. 2019 Feb;138(2):109-124. doi: 10.1007/s00439-019-01970-5. Epub 2019 Jan 22. Hum Genet. 2019. PMID: 30671672 Free PMC article. Review.
-
Protein-protein interaction extraction with feature selection by evaluating contribution levels of groups consisting of related features.BMC Bioinformatics. 2016 Jul 25;17 Suppl 7(Suppl 7):246. doi: 10.1186/s12859-016-1100-z. BMC Bioinformatics. 2016. PMID: 27454611 Free PMC article.
-
Automated recognition of functional compound-protein relationships in literature.PLoS One. 2020 Mar 3;15(3):e0220925. doi: 10.1371/journal.pone.0220925. eCollection 2020. PLoS One. 2020. PMID: 32126064 Free PMC article.
References
-
- Blaschke C, Andrade MA, Ouzounis C, Valencia A. Automatic extraction of biological information from scientific text: protein-protein interactions. Proc Int Conf Intell Syst Mol Biol. 1999;7:60–67. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

