Evolution at increased error rate leads to the coexistence of multiple adaptive pathways in an RNA virus
- PMID: 23323937
- PMCID: PMC3556134
- DOI: 10.1186/1471-2148-13-11
Evolution at increased error rate leads to the coexistence of multiple adaptive pathways in an RNA virus
Abstract
Background: When beneficial mutations present in different genomes spread simultaneously in an asexual population, their fixation can be delayed due to competition among them. This interference among mutations is mainly determined by the rate of beneficial mutations, which in turn depends on the population size, the total error rate, and the degree of adaptation of the population. RNA viruses, with their large population sizes and high error rates, are good candidates to present a great extent of interference. To test this hypothesis, in the current study we have investigated whether competition among beneficial mutations was responsible for the prolonged presence of polymorphisms in the mutant spectrum of an RNA virus, the bacteriophage Qβ, evolved during a large number of generations in the presence of the mutagenic nucleoside analogue 5-azacytidine.
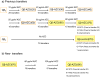
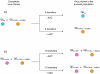
Results: The analysis of the mutant spectra of bacteriophage Qβ populations evolved at artificially increased error rate shows a large number of polymorphic mutations, some of them with demonstrated selective value. Polymorphisms distributed into several evolutionary lines that can compete among them, making it difficult the emergence of a defined consensus sequence. The presence of accompanying deleterious mutations, the high degree of recurrence of the polymorphic mutations, and the occurrence of epistatic interactions generate a highly complex interference dynamics.
Conclusions: Interference among beneficial mutations in bacteriophage Qβ evolved at increased error rate permits the coexistence of multiple adaptive pathways that can provide selective advantages by different molecular mechanisms. In this way, interference can be seen as a positive factor that allows the exploration of the different local maxima that exist in rugged fitness landscapes.
Figures

References
-
- Muller HJ. Some genetic aspects of sex. Am Nat. 1932;68:118–138.
-
- Muller HJ. The relation of recombination to mutational advance. Mutat Res. 1964;106:2–9. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

