A proteomics search algorithm specifically designed for high-resolution tandem mass spectra
- PMID: 23323968
- PMCID: PMC3586292
- DOI: 10.1021/pr301024c
A proteomics search algorithm specifically designed for high-resolution tandem mass spectra
Abstract
The acquisition of high-resolution tandem mass spectra (MS/MS) is becoming more prevalent in proteomics, but most researchers employ peptide identification algorithms that were designed prior to this development. Here, we demonstrate new software, Morpheus, designed specifically for high-mass accuracy data, based on a simple score that is little more than the number of matching products. For a diverse collection of data sets from a variety of organisms (E. coli, yeast, human) acquired on a variety of instruments (quadrupole-time-of-flight, ion trap-orbitrap, and quadrupole-orbitrap) in different laboratories, Morpheus gives more spectrum, peptide, and protein identifications at a 1% false discovery rate (FDR) than Mascot, Open Mass Spectrometry Search Algorithm (OMSSA), and Sequest. Additionally, Morpheus is 1.5 to 4.6 times faster, depending on the data set, than the next fastest algorithm, OMSSA. Morpheus was developed in C# .NET and is available free and open source under a permissive license.
Figures

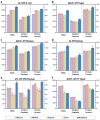
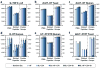

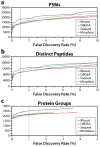
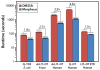
References
-
- Morris HR, Paxton T, Dell A, Langhorne J, Berg M, Bordoli RS, Hoyes J, Bateman RH. High sensitivity collisionally-activated decomposition tandem mass spectrometry on a novel quadrupole/orthogonal-acceleration time-of-flight mass spectrometer. Rapid Commun Mass Sp. 1996;10:889–896. - PubMed
-
- Kristensen DB, Imamura K, Miyamoto Y, Yoshizato K. Mass spectrometric approaches for the characterization of proteins on a hybrid quadrupole time-of-flight (Q-TOF) mass spectrometer. Electrophoresis. 2000;21:430–439. - PubMed
-
- Loboda AV, Krutchinsky AN, Bromirski M, Ens W, Standing KG. A tandem quadrupole/time-of-flight mass spectrometer with a matrix-assisted laser desorption/ionization source: design and performance. Rapid Commun Mass Sp. 2000;14:1047–1057. - PubMed
-
- Syka JEP, Marto JA, Bai DL, Horning S, Senko MW, Schwartz JC, Ueberheide B, Garcia B, Busby S, Muratore T, Shabanowitz J, Hunt DF. Novel linear quadrupole ion trap/FT mass spectrometer: Performance characterization and use in the comparative analysis of histone H3 post-translational modifications. J Proteome Res. 2004;3:621–626. - PubMed
-
- Makarov A, Denisov E, Kholomeev A, Baischun W, Lange O, Strupat K, Horning S. Performance evaluation of a hybrid linear ion trap/orbitrap mass spectrometer. Anal Chem. 2006;78:2113–2120. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

