Draft genome sequence of the male-killing Wolbachia strain wBol1 reveals recent horizontal gene transfers from diverse sources
- PMID: 23324387
- PMCID: PMC3639933
- DOI: 10.1186/1471-2164-14-20
Draft genome sequence of the male-killing Wolbachia strain wBol1 reveals recent horizontal gene transfers from diverse sources
Abstract
Background: The endosymbiont Wolbachia pipientis causes diverse and sometimes dramatic phenotypes in its invertebrate hosts. Four Wolbachia strains sequenced to date indicate that the constitution of the genome is dynamic, but these strains are quite divergent and do not allow resolution of genome diversification over shorter time periods. We have sequenced the genome of the strain wBol1-b, found in the butterfly Hypolimnas bolina, which kills the male offspring of infected hosts during embyronic development and is closely related to the non-male-killing strain wPip from Culex pipiens.
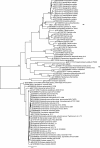
Results: The genomes of wBol1-b and wPip are similar in genomic organisation, sequence and gene content, but show substantial differences at some rapidly evolving regions of the genome, primarily associated with prophage and repetitive elements. We identified 44 genes in wBol1-b that do not have homologs in any previously sequenced strains, indicating that Wolbachia's non-core genome diversifies rapidly. These wBol1-b specific genes include a number that have been recently horizontally transferred from phylogenetically distant bacterial taxa. We further report a second possible case of horizontal gene transfer from a eukaryote into Wolbachia.
Conclusions: Our analyses support the developing view that many endosymbiotic genomes are highly dynamic, and are exposed and receptive to exogenous genetic material from a wide range of sources. These data also suggest either that this bacterial species is particularly permissive for eukaryote-to-prokaryote gene transfers, or that these transfers may be more common than previously believed. The wBol1-b-specific genes we have identified provide candidates for further investigations of the genomic bases of phenotypic differences between closely-related Wolbachia strains.
Figures


References
-
- Brownlie JC, Cass BN, Riegler M, Witsenburg JJ, Iturbe-Ormaetxe I, McGraw EA, O’Neill SL. Evidence for metabolic provisioning by a common invertebrate endosymbiont, Wolbachia pipientis, during periods of nutritional stress. PLoS Pathog. 2009;5(4):e1000368. doi: 10.1371/journal.ppat.1000368. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

