Escherichia coli- and Staphylococcus aureus-induced mastitis differentially modulate transcriptional responses in neighbouring uninfected bovine mammary gland quarters
- PMID: 23324411
- PMCID: PMC3598231
- DOI: 10.1186/1471-2164-14-36
Escherichia coli- and Staphylococcus aureus-induced mastitis differentially modulate transcriptional responses in neighbouring uninfected bovine mammary gland quarters
Abstract
Background: The most important disease of dairy cattle is mastitis, caused by the infection of the mammary gland by various micro-organisms. Although the transcriptional response of bovine mammary gland cells to in vitro infection has been studied, the interplay and consequences of these responses in the in vivo environment of the mammary gland are less clear. Previously mammary gland quarters were considered to be unaffected by events occurring in neighbouring quarters. More recently infection of individual quarters with mastitis causing pathogens, especially Escherichia coli, has been shown to influence the physiology of neighbouring uninfected quarters. Therefore, the transcriptional responses of uninfected mammary gland quarters adjacent to quarters infected with two major mastitis causing pathogens, E. coli and Staphylococcus aureus, were compared.
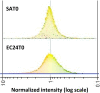
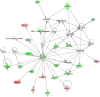
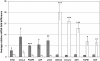
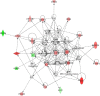
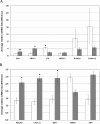
Results: The bacteriologically sterile, within-animal control quarters exhibited a transcriptional response to the infection of neighbouring quarters. The greatest response was associated with E. coli infection, while a weaker, yet significant, response occurred during S. aureus infection. The transcriptional responses of these uninfected quarters included the enhanced expression of many genes previously associated with mammary gland infections. Comparison of the transcriptional response of uninfected quarters to S. aureus and E. coli infection identified 187 differentially expressed genes, which were particularly associated with cellular responses, e.g. response to stress. The most affected network identified by Ingenuity Pathway analysis has the immunosuppressor transforming growth factor beta 1 (TGFB1) at its hub and largely consists of genes more highly expressed in control quarters from S. aureus infected cows.
Conclusions: Uninfected mammary gland quarters reacted to the infection of neighbouring quarters and the responses were dependent on pathogen type. Therefore, bovine udder quarters exhibit interdependence and should not be considered as separate functional entities. This suggests that mastitis pathogens not only interact directly with host mammary cells, but also influence discrete sites some distance away, which will affect their response to the subsequent spread of the infection. Understanding the underlying mechanisms may provide further clues for ways to control mammary gland infections. These results also have implications for the design of experimental studies investigating immune regulatory mechanisms in the bovine mammary gland.
Figures
References
-
- Viguier C, Arora S, Gilmartin N, Welbeck K, O’Kennedy R. Mastitis detection: current trends and future perspectives. Trends Biotech. 2009;27:486–493. - PubMed
-
- Schukken YH, Günther J, Fitzpatrick J, Fontaine MC, Goetze L, Holst O, Leigh J, Petzl W, Schuberth HJ, Sipka A, Smith DGE, Quesnell R, Watts J, Yancey R, Zerbe H, Gurjar A, Zadoks RN, Seyfert HM. Host-response patterns of intramammary infections in dairy cows. Vet Immunol Immunopath. 2011;144:270–289. - PubMed
-
- Rinaldi M, Li RW, Bannerman DD, Daniels KM, Evock-Clover C, Silva MVB, Paape MJ, Van Ryssen B, Burvenich C, Capuco AV. A sentinel function for teat tissues in dairy cows: dominant innate immune response elements define early response to E. coli mastitis. Funct Integr Genomics. 2010;10:21–38. - PubMed
-
- Moyes KM, Drackley JK, Morin DE, Bionaz M, Rodriguez-Zas SL, Everts RE, Lewin HA, Loor JJ. Gene network and pathway analysis of bovine mammary tissue challenged with Streptococcus uberis reveals induction of cell proliferation and inhibition of PPARγ signalling as potential mechanism for the negative relationships between immune response and lipid metabolism. BMC Genomics. 2009;10:542. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

