Comparative genome analysis of Spiroplasma melliferum IPMB4A, a honeybee-associated bacterium
- PMID: 23324436
- PMCID: PMC3563533
- DOI: 10.1186/1471-2164-14-22
Comparative genome analysis of Spiroplasma melliferum IPMB4A, a honeybee-associated bacterium
Abstract
Background: The genus Spiroplasma contains a group of helical, motile, and wall-less bacteria in the class Mollicutes. Similar to other members of this class, such as the animal-pathogenic Mycoplasma and the plant-pathogenic 'Candidatus Phytoplasma', all characterized Spiroplasma species were found to be associated with eukaryotic hosts. While most of the Spiroplasma species appeared to be harmless commensals of insects, a small number of species have evolved pathogenicity toward various arthropods and plants. In this study, we isolated a novel strain of honeybee-associated S. melliferum and investigated its genetic composition and evolutionary history by whole-genome shotgun sequencing and comparative analysis with other Mollicutes genomes.
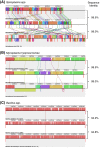
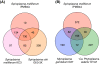
Results: The whole-genome shotgun sequencing of S. melliferum IPMB4A produced a draft assembly that was ~1.1 Mb in size and covered ~80% of the chromosome. Similar to other Spiroplasma genomes that have been studied to date, we found that this genome contains abundant repetitive sequences that originated from plectrovirus insertions. These phage fragments represented a major obstacle in obtaining a complete genome sequence of Spiroplasma with the current sequencing technology. Comparative analysis of S. melliferum IPMB4A with other Spiroplasma genomes revealed that these phages may have facilitated extensive genome rearrangements in these bacteria and contributed to horizontal gene transfers that led to species-specific adaptation to different eukaryotic hosts. In addition, comparison of gene content with other Mollicutes suggested that the common ancestor of the SEM (Spiroplasma, Entomoplasma, and Mycoplasma) clade may have had a relatively large genome and flexible metabolic capacity; the extremely reduced genomes of present day Mycoplasma and 'Candidatus Phytoplasma' species are likely to be the result of independent gene losses in these lineages.
Conclusions: The findings in this study highlighted the significance of phage insertions and horizontal gene transfer in the evolution of bacterial genomes and acquisition of pathogenicity. Furthermore, the inclusion of Spiroplasma in comparative analysis has improved our understanding of genome evolution in Mollicutes. Future improvements in the taxon sampling of available genome sequences in this group are required to provide further insights into the evolution of these important pathogens of humans, animals, and plants.
Figures




References
-
- Whitcomb RF. The biology of spiroplasmas. Ann. 1981;26:397–425. doi: 10.1146/annurev.en.26.010181.002145. - DOI
-
- Gasparich GE, Whitcomb RF, Dodge D, French FE, Glass J, Williamson DL. The genus Spiroplasma and its non-helical descendants: phylogenetic classification, correlation with phenotype and roots of the Mycoplasma mycoides clade. Int J Syst Evol Microbiol. 2004;54:893–918. doi: 10.1099/ijs.0.02688-0. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

