An R2R3 MYB transcription factor determines red petal colour in an Actinidia (kiwifruit) hybrid population
- PMID: 23324587
- PMCID: PMC3618344
- DOI: 10.1186/1471-2164-14-28
An R2R3 MYB transcription factor determines red petal colour in an Actinidia (kiwifruit) hybrid population
Abstract
Background: Red colour in kiwifruit results from the presence of anthocyanin pigments. Their expression, however, is complex, and varies among genotypes, species, tissues and environments. An understanding of the biosynthesis, physiology and genetics of the anthocyanins involved, and the control of their expression in different tissues, is required. A complex, the MBW complex, consisting of R2R3-MYB and bHLH transcription factors together with a WD-repeat protein, activates anthocyanin 3-O-galactosyltransferase (F3GT1) to produce anthocyanins. We examined the expression and genetic control of anthocyanins in flowers of Actinidia hybrid families segregating for red and white petal colour.
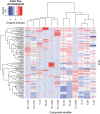
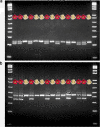
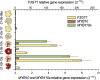
Results: Four inter-related backcross families between Actinidia chinensis Planch. var. chinensis and Actinidia eriantha Benth. were identified that segregated 1:1 for red or white petal colour. Flower pigments consisted of five known anthocyanins (two delphinidin-based and three cyanidin-based) and three unknowns. Intensity and hue differed in red petals from pale pink to deep magenta, and while intensity of colour increased with total concentration of anthocyanin, no association was found between any particular anthocyanin data and hue. Real time qPCR demonstrated that an R2R3 MYB, MYB110a, was expressed at significant levels in red-petalled progeny, but not in individuals with white petals.A microsatellite marker was developed that identified alleles that segregated with red petal colour, but not with ovary, stamen filament, or fruit flesh colour in these families. The marker mapped to chromosome 10 in Actinidia.The white petal phenotype was complemented by syringing Agrobacterium tumefaciens carrying Actinidia 35S::MYB110a into the petal tissue. Red pigments developed in white petals both with, and without, co-transformation with Actinidia bHLH partners. MYB110a was shown to directly activate Actinidia F3GT1 in transient assays.
Conclusions: The transcription factor, MYB110a, regulates anthocyanin production in petals in this hybrid population, but not in other flower tissues or mature fruit. The identification of delphinidin-based anthocyanins in these flowers provides candidates for colour enhancement in novel fruits.
Figures






References
-
- Gould K, Lister C. In: Flavonoids: Chemistry, Biochemistry, and Applications. Andersen OM, Markham K, editor. Boca Raton, Florida: CRC Press; 2006. Flavonoid functions in plants; pp. 397–411.
-
- de Pascual-Teresa S, Sanchez-Ballesta MT. Anthocyanins: from plant to health. Phytochem Rev. 2008;7:281–299. doi: 10.1007/s11101-007-9074-0. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

