A tissue-based comparative effectiveness analysis of biomarkers for early detection of colorectal tumors
- PMID: 23324654
- PMCID: PMC3535074
- DOI: 10.1038/ctg.2012.21
A tissue-based comparative effectiveness analysis of biomarkers for early detection of colorectal tumors
Abstract
Objectives: We recently identified a six-gene methylation-based biomarker panel suitable for early detection of colorectal cancer (CRC). In this study, we compared the performance of this novel epi-panel with that of previously identified DNA methylation markers in the same clinical tissue sample sets.
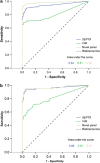
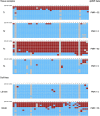
Methods: Quantitative methylation-specific PCR was used to analyze the promoter region of SEPT9 and VIM in a total of 485 tissue samples, divided into test and validation sets. ITGA4, NTRK2, OSMR, and TUBG2 were also included in the analyses. Receiver operating characteristic (ROC) curves were used to compare the performances of the individual biomarkers with that of the novel epi-panel.
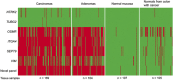
Results: SEPT9 and VIM were methylated in 82 and 67% of CRCs (n=169) and in 88 and 54% of the adenomas (n=104). Only 3% of the normal mucosa samples (n=107) were methylated for these genes, confirming that the methylation was highly cancer-specific. Areas under the ROC curve (AUC), distinguishing CRCs from normal mucosa, were 0.94 for SEPT9 and 0.81 for VIM. AUC values for separating adenomas from normal mucosa samples were 0.96 and 0.81 for the same genes. In comparison, the novel epi-panel achieved an AUC of 0.98 (CRC) and 0.97 (adenomas).ITGA4, OSMR, NTRK2, and TUBG2 were methylated in 90, 78, 7, and 1% of the CRCs, and in 76, 77, 3, and 0% of the adenomas. Between 0 and 2% of the normal mucosa samples were methylated for the same genes. ITGA4 and OSMR achieved an AUC of 0.96 and 0.92 (CRC vs. normal mucosa), and 0.93 and 0.92 (adenomas vs. normal mucosa).
Conclusions: We have confirmed the high performance of some of the previously identified DNA methylation markers. Furthermore, we showed that a recently reported epi-panel performed better than the individual DNA methylation biomarkers when analyzed in the same tissue samples. This observation was also true for VIM and SEPT9, which are included in commercially available noninvasive tests for CRC. These results further underscore the value of combining a manageable number of individual markers into a panel, which in addition to having a higher sensitivity and specificity might provide a more profound robustness to a noninvasive test compared with single markers.
Figures
Similar articles
-
Identification of an epigenetic biomarker panel with high sensitivity and specificity for colorectal cancer and adenomas.Mol Cancer. 2011 Jul 21;10:85. doi: 10.1186/1476-4598-10-85. Mol Cancer. 2011. PMID: 21777459 Free PMC article.
-
Combined detection of plasma GATA5 and SFRP2 methylation is a valid noninvasive biomarker for colorectal cancer and adenomas.World J Gastroenterol. 2015 Mar 7;21(9):2629-37. doi: 10.3748/wjg.v21.i9.2629. World J Gastroenterol. 2015. PMID: 25759530 Free PMC article.
-
Performance Comparison Between Plasma and Stool Methylated SEPT9 Tests for Detecting Colorectal Cancer.Front Genet. 2020 Apr 16;11:324. doi: 10.3389/fgene.2020.00324. eCollection 2020. Front Genet. 2020. PMID: 32373158 Free PMC article.
-
Status of integrin subunit alpha 4 promoter DNA methylation in colorectal cancer and other malignant tumors: a systematic review and meta-analysis.Res Pharm Sci. 2023 Mar 10;18(3):231-243. doi: 10.4103/1735-5362.371580. eCollection 2023 May-Jun. Res Pharm Sci. 2023. PMID: 37593168 Free PMC article. Review.
-
Advance in plasma SEPT9 gene methylation assay for colorectal cancer early detection.World J Gastrointest Oncol. 2018 Jan 15;10(1):15-22. doi: 10.4251/wjgo.v10.i1.15. World J Gastrointest Oncol. 2018. PMID: 29375744 Free PMC article. Review.
Cited by
-
Comparing the Cost-Effectiveness of Innovative Colorectal Cancer Screening Tests.J Natl Cancer Inst. 2021 Feb 1;113(2):154-161. doi: 10.1093/jnci/djaa103. J Natl Cancer Inst. 2021. PMID: 32761199 Free PMC article.
-
Candidate Molecular Biomarkers for the Non- Invasive Detection of Colorectal Cancer using Gene Expression Profiling.Iran J Pathol. 2021 Spring;16(2):205-214. doi: 10.30699/IJP.2021.132385.2475. Epub 2021 Mar 2. Iran J Pathol. 2021. PMID: 33936232 Free PMC article.
-
DNA Methylation Change Profiling of Colorectal Disease: Screening towards Clinical Use.Life (Basel). 2021 Apr 30;11(5):412. doi: 10.3390/life11050412. Life (Basel). 2021. PMID: 33946400 Free PMC article. Review.
-
The early predictive effect of low expression of the ITGA4 in colorectal cancer.J Gastrointest Oncol. 2022 Feb;13(1):265-278. doi: 10.21037/jgo-22-92. J Gastrointest Oncol. 2022. PMID: 35284127 Free PMC article.
-
Detection of aberrant methylated SEPT9 and NTRK3 genes in sporadic colorectal cancer patients as a potential diagnostic biomarker.Oncol Lett. 2016 Dec;12(6):5335-5343. doi: 10.3892/ol.2016.5327. Epub 2016 Oct 31. Oncol Lett. 2016. PMID: 28105243 Free PMC article.
References
-
- Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. - PubMed
-
- Atkin WS, Edwards R, Kralj-Hans I, et al. Once-only flexible sigmoidoscopy screening in prevention of colorectal cancer: a multicentre randomised controlled trial. Lancet. 2010;375:1624–1633. - PubMed
-
- Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143–153. - PubMed
-
- Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. - PubMed
-
- Chen WD, Han ZJ, Skoletsky J, et al. Detection in fecal DNA of colon cancer-specific methylation of the nonexpressed vimentin gene. J Natl Cancer Inst. 2005;97:1124–1132. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

