A computational approach to detect gap junction plaques and associate them with cells in fluorescent images
- PMID: 23324867
- PMCID: PMC3636683
- DOI: 10.1369/0022155413477114
A computational approach to detect gap junction plaques and associate them with cells in fluorescent images
Abstract
Intercellular signaling is a fundamental requirement for complex biological system function and survival. Communication between adjoining cells is largely achieved via gap junction channels made up of multiple subunits of connexin proteins, each with unique selectivity and regulatory properties. Intercellular communication via gap junction channels facilitates transmission of an array of cellular signals, including ions, macromolecules, and metabolites that coordinate physiological processes throughout tissues and entire organisms. Although current methods used to quantify connexin expression rely on number or area density measurements in a field of view, they lack cellular assignment, distance measurement capabilities (both within the cell and to extracellular structures), and complete automation. We devised an automated computational approach built on a contour expansion algorithm platform that allows connexin protein detection and assignment to specific cells within complex tissues. In addition, parallel implementation of the contour expansion algorithm allows for high-throughput analysis as the complexity of the biological sample increases. This method does not depend specifically on connexin identification and can be applied more widely to the analysis of numerous immunocytochemical markers as well as to identify particles within tissues such as nanoparticles, gene delivery vehicles, or even cellular fragments such as exosomes or microparticles.
Conflict of interest statement
Figures
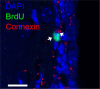
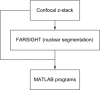
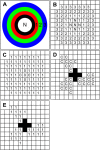

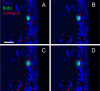

References
-
- Al-Kofahi Y, Lassoued W, Lee W, Royam B. 2010. Improved automatic detection and segmentation of cell nuclei in histopathology images. IEEE Trans Biomed Eng. 57:841–852 - PubMed
-
- Beyer EC. 1993. Gap junctions. Int Rev Cytol. 137C:1–37 - PubMed
-
- Cheng A, Tang H, Cai J, Zhu M, Zhang X, Rao M, Mattson MP. 2004. Gap junctional communication is required to maintain mouse cortical neural progenitor cells in a proliferative state. Dev Biol. 272:203–216 - PubMed
-
- Dang X, Doble BW, Kardami E. 2003. The carboxy-tail of connexin-43 localizes to the nucleus and inhibits cell growth. Mol Cell Biochem. 242(1–2):35–38 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

