Regulation of intestinal microbiota by the NLR protein family
- PMID: 23325116
- PMCID: PMC3597849
- DOI: 10.1093/intimm/dxs116
Regulation of intestinal microbiota by the NLR protein family
Abstract
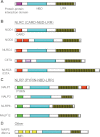
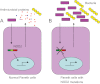
The human intestine harbors a diverse microbial community consisting of a large number of bacteria and other micro-organisms that have co-evolved with the host intestinal immune system. During this process, microbiota and the host immune system shape one another by various mechanisms to achieve a successful symbiotic relationship. An increasing amount of evidence suggests that dysbiosis--the breakdown of such harmonized colonization--may result in infectious and inflammatory disorders, and recent advances in our studies indicate that receptors such as Toll-like receptors and NLR (nucleotide-binding oligomerization domain-like receptor; or nucleotide-binding domain- and leucine-rich repeat-containing receptor) proteins that detect micro-organisms and their products play a critical role in maintaining intestinal homeostasis. In this review, we summarize the role of NLR proteins in the regulation of intestinal microbiota. NLR proteins belong to a diverse family of cytoplasmic microbial sensors, mutations of which are involved in various disorders, including inflammatory bowel diseases. Understanding of the different roles of NLR family proteins in the intestine is, therefore, an important step towards the development of therapeutics against digestive diseases.
Figures

Similar articles
-
Nod2: a key regulator linking microbiota to intestinal mucosal immunity.J Mol Med (Berl). 2012 Jan;90(1):15-24. doi: 10.1007/s00109-011-0802-y. Epub 2011 Aug 23. J Mol Med (Berl). 2012. PMID: 21861185 Free PMC article. Review.
-
Stories of love and hate: innate immunity and host-microbe crosstalk in the intestine.Curr Opin Gastroenterol. 2013 Mar;29(2):125-32. doi: 10.1097/MOG.0b013e32835da2c7. Curr Opin Gastroenterol. 2013. PMID: 23337934 Review.
-
Cellular and molecular mechanisms underlying NOD2 risk-associated polymorphisms in Crohn's disease.Immunol Rev. 2014 Jul;260(1):249-60. doi: 10.1111/imr.12193. Immunol Rev. 2014. PMID: 24942694 Review.
-
Nucleotide-binding-oligomerization domain proteins and toll-like receptors: sensors of the inflammatory bowel diseases' microbial environment.Curr Opin Gastroenterol. 2005 Jul;21(4):419-25. Curr Opin Gastroenterol. 2005. PMID: 15930981 Review.
-
Interactions between the host innate immune system and microbes in inflammatory bowel disease.Gastroenterology. 2011 May;140(6):1729-37. doi: 10.1053/j.gastro.2011.02.012. Gastroenterology. 2011. PMID: 21530739 Free PMC article. Review.
Cited by
-
Detection of enteric pathogens by the nodosome.Trends Immunol. 2014 Mar;35(3):123-30. doi: 10.1016/j.it.2013.10.009. Epub 2013 Nov 22. Trends Immunol. 2014. PMID: 24268520 Free PMC article. Review.
-
Host genetics and diet, but not immunoglobulin A expression, converge to shape compositional features of the gut microbiome in an advanced intercross population of mice.Genome Biol. 2014;15(12):552. doi: 10.1186/s13059-014-0552-6. Genome Biol. 2014. PMID: 25516416 Free PMC article.
-
Bifidobacterium longum CCM 7952 Promotes Epithelial Barrier Function and Prevents Acute DSS-Induced Colitis in Strictly Strain-Specific Manner.PLoS One. 2015 Jul 28;10(7):e0134050. doi: 10.1371/journal.pone.0134050. eCollection 2015. PLoS One. 2015. PMID: 26218526 Free PMC article.
-
Intestinal microbiota: a new force in cancer immunotherapy.Cell Commun Signal. 2020 Jun 10;18(1):90. doi: 10.1186/s12964-020-00599-6. Cell Commun Signal. 2020. PMID: 32522267 Free PMC article. Review.
-
Dietary input of microbes and host genetic variation shape among-population differences in stickleback gut microbiota.ISME J. 2015 Nov;9(11):2515-26. doi: 10.1038/ismej.2015.64. Epub 2015 Apr 24. ISME J. 2015. PMID: 25909977 Free PMC article.
References
-
- Hooper L. V., Gordon J. I. 2001. Commensal host-bacterial relationships in the gut. Science 292: 1115 - PubMed
-
- Guarner F., Malagelada J. R. 2003. Gut flora in health and disease. Lancet 361: 512 - PubMed
-
- Xavier R. J., Podolsky D. K. 2007. Unravelling the pathogenesis of inflammatory bowel disease. Nature 448: 427 - PubMed
-
- Sartor R. B. 2008. Microbial influences in inflammatory bowel diseases. Gastroenterology 134: 577 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

