PPInterFinder--a mining tool for extracting causal relations on human proteins from literature
- PMID: 23325628
- PMCID: PMC3548331
- DOI: 10.1093/database/bas052
PPInterFinder--a mining tool for extracting causal relations on human proteins from literature
Abstract
One of the most common and challenging problem in biomedical text mining is to mine protein-protein interactions (PPIs) from MEDLINE abstracts and full-text research articles because PPIs play a major role in understanding the various biological processes and the impact of proteins in diseases. We implemented, PPInterFinder--a web-based text mining tool to extract human PPIs from biomedical literature. PPInterFinder uses relation keyword co-occurrences with protein names to extract information on PPIs from MEDLINE abstracts and consists of three phases. First, it identifies the relation keyword using a parser with Tregex and a relation keyword dictionary. Next, it automatically identifies the candidate PPI pairs with a set of rules related to PPI recognition. Finally, it extracts the relations by matching the sentence with a set of 11 specific patterns based on the syntactic nature of PPI pair. We find that PPInterFinder is capable of predicting PPIs with the accuracy of 66.05% on AIMED corpus and outperforms most of the existing systems. DATABASE URL: http://www.biomining-bu.in/ppinterfinder/
Figures


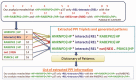


Similar articles
-
Automated Extraction and Visualization of Protein-Protein Interaction Networks and Beyond: A Text-Mining Protocol.Methods Mol Biol. 2020;2074:13-34. doi: 10.1007/978-1-4939-9873-9_2. Methods Mol Biol. 2020. PMID: 31583627
-
PPI finder: a mining tool for human protein-protein interactions.PLoS One. 2009;4(2):e4554. doi: 10.1371/journal.pone.0004554. Epub 2009 Feb 23. PLoS One. 2009. PMID: 19234603 Free PMC article.
-
Towards Extracting Supporting Information About Predicted Protein-Protein Interactions.IEEE/ACM Trans Comput Biol Bioinform. 2018 Jul-Aug;15(4):1239-1246. doi: 10.1109/TCBB.2015.2505278. Epub 2015 Dec 7. IEEE/ACM Trans Comput Biol Bioinform. 2018. PMID: 26672046
-
Mining biological networks from full-text articles.Methods Mol Biol. 2014;1159:135-45. doi: 10.1007/978-1-4939-0709-0_8. Methods Mol Biol. 2014. PMID: 24788265 Review.
-
Event extraction for systems biology by text mining the literature.Trends Biotechnol. 2010 Jul;28(7):381-90. doi: 10.1016/j.tibtech.2010.04.005. Epub 2010 Jun 1. Trends Biotechnol. 2010. PMID: 20570001 Review.
Cited by
-
An overview of the BioCreative 2012 Workshop Track III: interactive text mining task.Database (Oxford). 2013 Jan 17;2013:bas056. doi: 10.1093/database/bas056. Print 2013. Database (Oxford). 2013. PMID: 23327936 Free PMC article.
-
Large Language Models and Genomics for Summarizing the Role of microRNA in Regulating mRNA Expression.Biomedicines. 2024 Jul 10;12(7):1535. doi: 10.3390/biomedicines12071535. Biomedicines. 2024. PMID: 39062108 Free PMC article.
-
Reduction of supervision for biomedical knowledge discovery.BMC Bioinformatics. 2025 Sep 1;26(1):225. doi: 10.1186/s12859-025-06187-0. BMC Bioinformatics. 2025. PMID: 40890616 Free PMC article.
-
A comprehensive large scale biomedical knowledge graph for AI powered data driven biomedical research.bioRxiv [Preprint]. 2025 Mar 4:2023.10.13.562216. doi: 10.1101/2023.10.13.562216. bioRxiv. 2025. PMID: 38168218 Free PMC article. Preprint.
-
Towards Evidence-based Precision Medicine: Extracting Population Information from Biomedical Text using Binary Classifiers and Syntactic Patterns.AMIA Jt Summits Transl Sci Proc. 2016 Jul 20;2016:203-12. eCollection 2016. AMIA Jt Summits Transl Sci Proc. 2016. PMID: 27570671 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

