Comparative proteomic analysis of HIV-1 particles reveals a role for Ezrin and EHD4 in the Nef-dependent increase of virus infectivity
- PMID: 23325686
- PMCID: PMC3624205
- DOI: 10.1128/JVI.02477-12
Comparative proteomic analysis of HIV-1 particles reveals a role for Ezrin and EHD4 in the Nef-dependent increase of virus infectivity
Abstract
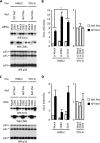
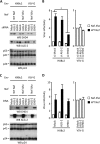
Nef is a human immunodeficiency virus type 1 (HIV-1) auxiliary protein that plays an important role in virus replication and the onset of acquired immunodeficiency. Although known functions of Nef might explain its contribution to HIV-1-associated pathogenesis, how Nef increases virus infectivity is still an open question. In vitro, Nef-deleted viruses have a defect that prevents efficient completion of early steps of replication. We have previously shown that this restriction is not due to the absence of Nef in viral particles. Rather, a loss of function in virus-producing cells accounts for the lower infectivity of nef-deleted viruses compared to wild-type (WT) viruses. Here we used DiGE and iTRAQ to identify differences between the proteomes of WT and nef-deleted viruses. We observe that glucosidase II is enriched in WT virions, whereas Ezrin, ALG-2, CD81, and EHD4 are enriched in nef-deleted virions. Functional analysis shows that glucosidase II, ALG-2, and CD81 have no or only Nef-independent effect on infectivity. In contrast, Ezrin and EHD4 are involved in the ability of Nef to increase virus infectivity (referred to thereafter as Nef potency). Indeed, simultaneous Ezrin and EHD4 depletion in SupT1 and 293T virus-producing cells result in an ∼30 and ∼70% decrease of Nef potency, respectively. Finally, while Ezrin behaves as an inhibitory factor counteracted by Nef, EHD4 should be considered as a cofactors required by Nef to increase virus infectivity.
Figures




References
-
- Sheehy AM, Gaddis NC, Choi JD, Malim MH. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646–650 - PubMed
-
- Neil SJ, Zang T, Bieniasz PD. 2008. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451:425–430 - PubMed
-
- Garcia JV, Miller AD. 1991. Serine phosphorylation-independent downregulation of cell-surface CD4 by nef. Nature 350:508–511 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

