Impact of CCR2 and SDF1 polymorphisms on disease progression in HIV-infected subjects in Thailand
- PMID: 23325742
- PMCID: PMC6807585
- DOI: 10.1002/jcla.21559
Impact of CCR2 and SDF1 polymorphisms on disease progression in HIV-infected subjects in Thailand
Abstract
Background: The genotypic polymorphisms of CCR5, CCR2, and SDF1 were analyzed to determine their impact as potential confounders with regard to disease progression because of the role that host genetic factors appear to be involved in determining rates of disease progression.
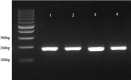
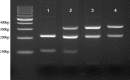
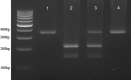
Methods: Genomic DNA was extracted from Ethylenediaminetetraacetate whole blood using Qiagen DNA extraction kit. The amplification of CCR5, CCR2, and SDF1 genes was performed by PCR.
Results: Two hundred and twenty-one samples were genotyped for the CCR5, CCR2, and SDF1 mutation. Among these, all (100%) were identified as wild type for CCR5. All were then investigated considering the impact on CD4+ T-cell counts. Samples were divided into two groups based on the CD4+ T-cell numbers. It revealed that in the group of CD4+ T-cell counts ≥200 cells/μl, 15 were found for the homozygous for SDF1 gene (3'A/3'A) whereas one was found in the group of CD4+ T-cell counts <200 cells/μl. Homozygosity for the CCR2 polymorphisms (64I/64I) were five in the group of CD4+ T-cell counts ≥200 cells/μl and none were found in the group of CD4+ T-cell counts <200 cells/μl. These results demonstrated that there was a significant association between CD4+ T-cell numbers and CCR2 and SDF1 polymorphisms (P < 0.001).
Conclusions: The mutation of CCR2 and SDF1 genes showed a significant difference in the distribution of CD4+ T-cell numbers (P < 0.001) whereas mutation of chemokine coreceptor CCR5 was not appeared to be associated with the impact of CD4+ T-cell counts.
© 2012 Wiley Periodicals, Inc.
Figures
References
-
- Smith MW, Dean M, Carrington M, et al. Contrasting genetic influence of CCR2 and CCR5 variants on HIV‐1 infection and disease progression. Hemophilia Growth and Development Study (HGDS), Multicenter AIDS Cohort Study (MACS), Multicenter Hemophilia Cohort Study (MHCS), San Francisco City Cohort (SFCC), ALIVE Study. Science 1997;277:959–965. - PubMed
-
- Xu L, Qiao Y, Zhang X, et al. CCR2–64I allele is associated with the progression of AIDS in a Han Chinese population. Mol Biol Rep 2010;37:311–326. - PubMed
-
- Ma L, Marmor M, Zhong P, Ewane L, Su B, Nyambi P. Distribution of CCR2–64I and SDF1–3′A alleles and HIV status in 7 ethnic populations of Cameroon. J Acquir Immune Defic Syndr 2005;40:89–95. - PubMed
-
- Tan XH, Zhang JY, Di CH, et al. Distribution of CCR5‐Delta32, CCR5m303A, CCR2–64I and SDF1–3′A in HIV‐1 infected and uninfected high‐risk Uighurs in Xinjiang, China. Infect Genet Evol 2010;10:268–272. - PubMed
-
- Salem AH, Farid E, Fadel R, et al. Distribution of four HIV type 1‐resistance polymorphisms (CCR5‐Delta32, CCR5‐m303, CCR2–64I, and SDF1–3′A) in the Bahraini population. AIDS Res Hum Retroviruses 2009;25:973–977. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

