Glycosaminoglycan glycomics using mass spectrometry
- PMID: 23325770
- PMCID: PMC3617335
- DOI: 10.1074/mcp.R112.026294
Glycosaminoglycan glycomics using mass spectrometry
Abstract
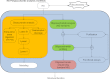
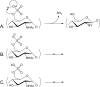
The fact that sulfated glycosaminoglycans (GAGs) are necessary for the functioning of all animal physiological systems drives the need to understand their biology. This understanding is limited, however, by the heterogeneous nature of GAG chains and their dynamic spatial and temporal expression patterns. GAGs have a regulated structure overlaid by heterogeneity but lack the detail necessary to build structure/function relationships. In order to provide this information, we need glycomics platforms that are sensitive, robust, high throughput, and information rich. This review summarizes progress on mass-spectrometry-based GAG glycomics methods. The areas covered include disaccharide analysis, oligosaccharide profiling, and tandem mass spectrometric sequencing.
Figures


Similar articles
-
On-line separations combined with MS for analysis of glycosaminoglycans.Mass Spectrom Rev. 2009 Mar-Apr;28(2):254-72. doi: 10.1002/mas.20200. Mass Spectrom Rev. 2009. PMID: 18956477 Free PMC article. Review.
-
Glycomics approach to structure-function relationships of glycosaminoglycans.Annu Rev Biomed Eng. 2006;8:181-231. doi: 10.1146/annurev.bioeng.8.061505.095745. Annu Rev Biomed Eng. 2006. PMID: 16834555 Review.
-
Glycosaminoglycan characterization by electrospray ionization mass spectrometry including fourier transform mass spectrometry.Methods Enzymol. 2010;478:79-108. doi: 10.1016/S0076-6879(10)78003-4. Methods Enzymol. 2010. PMID: 20816475 Free PMC article.
-
Proteoglycomics: recent progress and future challenges.OMICS. 2010 Aug;14(4):389-99. doi: 10.1089/omi.2009.0123. OMICS. 2010. PMID: 20450439 Free PMC article. Review.
-
Recent advances in glycosaminoglycan analysis by various mass spectrometry techniques.Anal Bioanal Chem. 2019 Jul;411(17):3731-3741. doi: 10.1007/s00216-019-01722-4. Epub 2019 Mar 26. Anal Bioanal Chem. 2019. PMID: 30911798 Review.
Cited by
-
Improved de novo sequencing of heparin/heparan sulfate oligosaccharides by propionylation of sites of sulfation.Carbohydr Res. 2018 Jul 30;465:16-21. doi: 10.1016/j.carres.2018.06.002. Epub 2018 Jun 8. Carbohydr Res. 2018. PMID: 29920400 Free PMC article.
-
A computational framework for heparan sulfate sequencing using high-resolution tandem mass spectra.Mol Cell Proteomics. 2014 Sep;13(9):2490-502. doi: 10.1074/mcp.M114.039560. Epub 2014 Jun 12. Mol Cell Proteomics. 2014. PMID: 24925905 Free PMC article.
-
GAG-DB, the New Interface of the Three-Dimensional Landscape of Glycosaminoglycans.Biomolecules. 2020 Dec 11;10(12):1660. doi: 10.3390/biom10121660. Biomolecules. 2020. PMID: 33322545 Free PMC article.
-
Online Capillary Zone Electrophoresis Negative Electron Transfer Dissociation Tandem Mass Spectrometry of Glycosaminoglycan Mixtures.Int J Mass Spectrom. 2019 Nov;445:116209. doi: 10.1016/j.ijms.2019.116209. Epub 2019 Aug 17. Int J Mass Spectrom. 2019. PMID: 32641905 Free PMC article.
-
Platelet Factor 4 Interactions with Short Heparin Oligomers: Implications for Folding and Assembly.Biophys J. 2020 Oct 6;119(7):1371-1379. doi: 10.1016/j.bpj.2020.04.012. Epub 2020 Apr 21. Biophys J. 2020. PMID: 32348723 Free PMC article.
References
-
- DeAngelis P. L. (2002) Evolution of glycosaminoglycans and their glycosyltransferases: implications for the extracellular matrices of animals and the capsules of pathogenic bacteria. Anat. Rec. 268, 317–326 - PubMed
-
- Bishop J. R., Schuksz M., Esko J. D. (2007) Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature 446, 1030–1037 - PubMed
-
- Bulow H. E., Hobert O. (2006) The molecular diversity of glycosaminoglycans shapes animal development. Annu. Rev. Cell Dev. Biol. 22, 375–407 - PubMed
-
- Chu C. L., Goerges A. L., Nugent M. A. (2005) Identification of common and specific growth factor binding sites in heparan sulfate proteoglycans. Biochemistry 44, 12203–12213 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

