Structural basis for a bispecific NADP+ and CoA binding site in an archaeal malonyl-coenzyme A reductase
- PMID: 23325803
- PMCID: PMC3585071
- DOI: 10.1074/jbc.M112.421263
Structural basis for a bispecific NADP+ and CoA binding site in an archaeal malonyl-coenzyme A reductase
Abstract
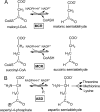
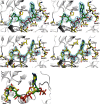
Autotrophic members of the Sulfolobales (crenarchaeota) use the 3-hydroxypropionate/4-hydroxybutyrate cycle to assimilate CO2 into cell material. The product of the initial acetyl-CoA carboxylation with CO2, malonyl-CoA, is further reduced to malonic semialdehyde by an NADPH-dependent malonyl-CoA reductase (MCR); the enzyme also catalyzes the reduction of succinyl-CoA to succinic semialdehyde onwards in the cycle. Here, we present the crystal structure of Sulfolobus tokodaii malonyl-CoA reductase in the substrate-free state and in complex with NADP(+) and CoA. Structural analysis revealed an unexpected reaction cycle in which NADP(+) and CoA successively occupy identical binding sites. Both coenzymes are pressed into an S-shaped, nearly superimposable structure imposed by a fixed and preformed binding site. The template-governed cofactor shaping implicates the same binding site for the 3'- and 2'-ribose phosphate group of CoA and NADP(+), respectively, but a different one for the common ADP part: the β-phosphate of CoA aligns with the α-phosphate of NADP(+). Evolution from an NADP(+) to a bispecific NADP(+) and CoA binding site involves many amino acid exchanges within a complex process by which constraints of the CoA structure also influence NADP(+) binding. Based on the paralogous aspartate-β-semialdehyde dehydrogenase structurally characterized with a covalent Cys-aspartyl adduct, a malonyl/succinyl group can be reliably modeled into MCR and discussed regarding its binding mode, the malonyl/succinyl specificity, and the catalyzed reaction. The modified polypeptide surrounding around the absent ammonium group in malonate/succinate compared with aspartate provides the structural basis for engineering a methylmalonyl-CoA reductase applied for biotechnical polyester building block synthesis.
Figures



Similar articles
-
Malonyl-coenzyme A reductase in the modified 3-hydroxypropionate cycle for autotrophic carbon fixation in archaeal Metallosphaera and Sulfolobus spp.J Bacteriol. 2006 Dec;188(24):8551-9. doi: 10.1128/JB.00987-06. Epub 2006 Oct 13. J Bacteriol. 2006. PMID: 17041055 Free PMC article.
-
Malonic semialdehyde reductase, succinic semialdehyde reductase, and succinyl-coenzyme A reductase from Metallosphaera sedula: enzymes of the autotrophic 3-hydroxypropionate/4-hydroxybutyrate cycle in Sulfolobales.J Bacteriol. 2009 Oct;191(20):6352-62. doi: 10.1128/JB.00794-09. Epub 2009 Aug 14. J Bacteriol. 2009. PMID: 19684143 Free PMC article.
-
Structural basis of a bi-functional malonyl-CoA reductase (MCR) from the photosynthetic green non-sulfur bacterium Roseiflexus castenholzii.mBio. 2023 Aug 31;14(4):e0323322. doi: 10.1128/mbio.03233-22. Epub 2023 Jun 6. mBio. 2023. PMID: 37278533 Free PMC article.
-
Malonyl-coenzyme A reductase from Chloroflexus aurantiacus, a key enzyme of the 3-hydroxypropionate cycle for autotrophic CO(2) fixation.J Bacteriol. 2002 May;184(9):2404-10. doi: 10.1128/JB.184.9.2404-2410.2002. J Bacteriol. 2002. PMID: 11948153 Free PMC article.
-
An evolutionary history of the CoA-binding protein Nat/Ivy.Protein Sci. 2022 Dec;31(12):e4463. doi: 10.1002/pro.4463. Protein Sci. 2022. PMID: 36192822 Free PMC article. Review.
Cited by
-
Crystal Structure of the LysY·LysW Complex from Thermus thermophilus.J Biol Chem. 2016 May 6;291(19):9948-59. doi: 10.1074/jbc.M115.707034. Epub 2016 Mar 9. J Biol Chem. 2016. PMID: 26966182 Free PMC article.
-
The aldehyde dehydrogenase superfamilies: correlations and deviations in structure and function.Appl Microbiol Biotechnol. 2025 Apr 29;109(1):106. doi: 10.1007/s00253-025-13467-5. Appl Microbiol Biotechnol. 2025. PMID: 40301148 Free PMC article. Review.
-
Lambda-PCR for precise DNA assembly and modification.Biotechnol Bioeng. 2022 Dec;119(12):3657-3667. doi: 10.1002/bit.28240. Epub 2022 Oct 8. Biotechnol Bioeng. 2022. PMID: 36148504 Free PMC article.
-
Structure of aspartate β-semialdehyde dehydrogenase from Francisella tularensis.Acta Crystallogr F Struct Biol Commun. 2018 Jan 1;74(Pt 1):14-22. doi: 10.1107/S2053230X17017241. Epub 2018 Jan 1. Acta Crystallogr F Struct Biol Commun. 2018. PMID: 29372903 Free PMC article.
-
Functional analysis of Rossmann-like domains reveals convergent evolution of topology and reaction pathways.PLoS Comput Biol. 2019 Dec 23;15(12):e1007569. doi: 10.1371/journal.pcbi.1007569. eCollection 2019 Dec. PLoS Comput Biol. 2019. PMID: 31869345 Free PMC article.
References
-
- Burton N. P., Williams T. D., Norris P. R. (1999) Carboxylase genes of Sulfolobus metallicus. Arch. Microbiol. 172, 349–353 - PubMed
-
- Norris P., Nixon A., Hart A. (1989) Acidophilic, mineral-oxidizing bacteria: the utilization of carbon dioxide with particular reference to autotrophy in Sulfolobus in Microbiology of Extreme Environments and Its Potential for Biotechnology (Da Costa M. S., Duarte J. C., Williams R. A. D., eds), pp. 24–39, Elsevier, London, United Kingdom: ) Elsevier, London, United Kingdom
-
- Menendez C., Bauer Z., Huber H., Gad'on N., Stetter K. O., Fuchs G. (1999) Presence of acetyl coenzyme A (CoA) carboxylase and propionyl-CoA carboxylase in autotrophic Crenarchaeota and indication for operation of a 3-hydroxypropionate cycle in autotrophic carbon fixation. J. Bacteriol. 181, 1088–1098 - PMC - PubMed
-
- Ishii M., Miyake T., Satoh T., Sugiyama H., Oshima Y., Kodama T., Igarashi Y. (1996) Autotrophic carbon dioxide fixation in Acidianus brierleyi. Arch. Microbiol. 166, 368–371 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

