Molecular characterization of high-level-cholera-toxin-producing El Tor variant Vibrio cholerae strains in the Zanzibar Archipelago of Tanzania
- PMID: 23325815
- PMCID: PMC3592071
- DOI: 10.1128/JCM.03162-12
Molecular characterization of high-level-cholera-toxin-producing El Tor variant Vibrio cholerae strains in the Zanzibar Archipelago of Tanzania
Abstract
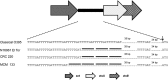
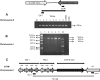
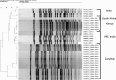
Analysis of 1,180 diarrheal stool samples in Zanzibar detected 247 Vibrio cholerae O1, Ogawa strains in 2009. Phenotypic traits and PCR-based detection of rstR, rtxC, and tcpA alleles showed that they belonged to the El Tor biotype. Genetic analysis of ctxB of these strains revealed that they were classical type, and production of classical cholera toxin B (CTB) was confirmed by Western blotting. These strains produced more CT than the prototype El Tor and formed a separate cluster by pulsed-field gel electrophoresis (PFGE) analysis.
Figures


References
-
- World Health Organization 2012. Cholera, 2012. Wkly. Epidemiol. Rec. 87:289–304
-
- Mintz ED, Guerrant RL. 2009. A lion in our village—the unconscionable tragedy of cholera in Africa. N. Engl. J. Med. 360:1060–1063 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

