Histone methyltransferase SETD2 coordinates FACT recruitment with nucleosome dynamics during transcription
- PMID: 23325844
- PMCID: PMC3597667
- DOI: 10.1093/nar/gks1472
Histone methyltransferase SETD2 coordinates FACT recruitment with nucleosome dynamics during transcription
Abstract
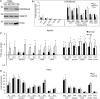
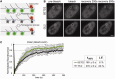
Histone H3 of nucleosomes positioned on active genes is trimethylated at Lys36 (H3K36me3) by the SETD2 (also termed KMT3A/SET2 or HYPB) methyltransferase. Previous studies in yeast indicated that H3K36me3 prevents spurious intragenic transcription initiation through recruitment of a histone deacetylase complex, a mechanism that is not conserved in mammals. Here, we report that downregulation of SETD2 in human cells leads to intragenic transcription initiation in at least 11% of active genes. Reduction of SETD2 prevents normal loading of the FACT (FAcilitates Chromatin Transcription) complex subunits SPT16 and SSRP1, and decreases nucleosome occupancy in active genes. Moreover, co-immunoprecipitation experiments suggest that SPT16 is recruited to active chromatin templates, which contain H3K36me3-modified nucleosomes. Our results further show that within minutes after transcriptional activation, there is a SETD2-dependent reduction in gene body occupancy of histone H2B, but not of histone H3, suggesting that SETD2 coordinates FACT-mediated exchange of histone H2B during transcription-coupled nucleosome displacement. After inhibition of transcription, we observe a SETD2-dependent recruitment of FACT and increased histone H2B occupancy. These data suggest that SETD2 activity modulates FACT recruitment and nucleosome dynamics, thereby repressing cryptic transcription initiation.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

