A guide to enterotypes across the human body: meta-analysis of microbial community structures in human microbiome datasets
- PMID: 23326225
- PMCID: PMC3542080
- DOI: 10.1371/journal.pcbi.1002863
A guide to enterotypes across the human body: meta-analysis of microbial community structures in human microbiome datasets
Abstract
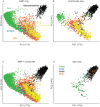
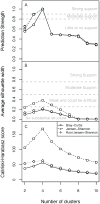
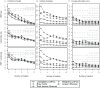
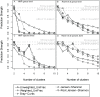
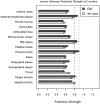
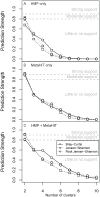
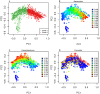
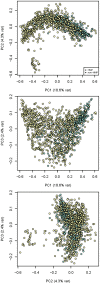
Recent analyses of human-associated bacterial diversity have categorized individuals into 'enterotypes' or clusters based on the abundances of key bacterial genera in the gut microbiota. There is a lack of consensus, however, on the analytical basis for enterotypes and on the interpretation of these results. We tested how the following factors influenced the detection of enterotypes: clustering methodology, distance metrics, OTU-picking approaches, sequencing depth, data type (whole genome shotgun (WGS) vs.16S rRNA gene sequence data), and 16S rRNA region. We included 16S rRNA gene sequences from the Human Microbiome Project (HMP) and from 16 additional studies and WGS sequences from the HMP and MetaHIT. In most body sites, we observed smooth abundance gradients of key genera without discrete clustering of samples. Some body habitats displayed bimodal (e.g., gut) or multimodal (e.g., vagina) distributions of sample abundances, but not all clustering methods and workflows accurately highlight such clusters. Because identifying enterotypes in datasets depends not only on the structure of the data but is also sensitive to the methods applied to identifying clustering strength, we recommend that multiple approaches be used and compared when testing for enterotypes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

