Assembly of the type II secretion system such as found in Vibrio cholerae depends on the novel Pilotin AspS
- PMID: 23326233
- PMCID: PMC3542185
- DOI: 10.1371/journal.ppat.1003117
Assembly of the type II secretion system such as found in Vibrio cholerae depends on the novel Pilotin AspS
Abstract
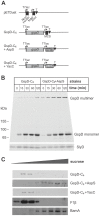
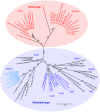
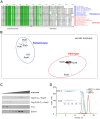
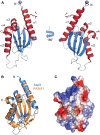
The Type II Secretion System (T2SS) is a molecular machine that drives the secretion of fully-folded protein substrates across the bacterial outer membrane. A key element in the machinery is the secretin: an integral, multimeric outer membrane protein that forms the secretion pore. We show that three distinct forms of T2SSs can be distinguished based on the sequence characteristics of their secretin pores. Detailed comparative analysis of two of these, the Klebsiella-type and Vibrio-type, showed them to be further distinguished by the pilotin that mediates their transport and assembly into the outer membrane. We have determined the crystal structure of the novel pilotin AspS from Vibrio cholerae, demonstrating convergent evolution wherein AspS is functionally equivalent and yet structurally unrelated to the pilotins found in Klebsiella and other bacteria. AspS binds to a specific targeting sequence in the Vibrio-type secretins, enhances the kinetics of secretin assembly, and homologs of AspS are found in all species of Vibrio as well those few strains of Escherichia and Shigella that have acquired a Vibrio-type T2SS.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Bos MP, Robert V, Tommassen J (2007) Biogenesis of the gram-negative bacterial outer membrane. Annu Rev Microbiol 61: 191–214. - PubMed
-
- Knowles TJ, Scott-Tucker A, Overduin M, Henderson IR (2009) Membrane protein architects: the role of the BAM complex in outer membrane protein assembly. Nat Rev Microbiol 7: 206–214. - PubMed
-
- Hagan CL, Silhavy TJ, Kahne D (2011) beta-Barrel membrane protein assembly by the Bam complex. Annu Rev Biochem 80: 189–210. - PubMed
-
- Tokuda H, Matsuyama S (2004) Sorting of lipoproteins to the outer membrane in E. coli. Biochim Biophys Acta 1693: 5–13. - PubMed
-
- Okuda S, Tokuda H (2011) Lipoprotein sorting in bacteria. Annu Rev Microbiol 65: 239–259. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

