Increased maternal genome dosage bypasses the requirement of the FIS polycomb repressive complex 2 in Arabidopsis seed development
- PMID: 23326241
- PMCID: PMC3542072
- DOI: 10.1371/journal.pgen.1003163
Increased maternal genome dosage bypasses the requirement of the FIS polycomb repressive complex 2 in Arabidopsis seed development
Abstract
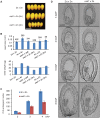
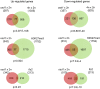
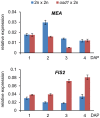
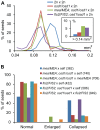
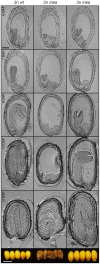
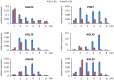
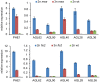
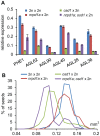
Seed development in flowering plants is initiated after a double fertilization event with two sperm cells fertilizing two female gametes, the egg cell and the central cell, leading to the formation of embryo and endosperm, respectively. In most species the endosperm is a polyploid tissue inheriting two maternal genomes and one paternal genome. As a consequence of this particular genomic configuration the endosperm is a dosage sensitive tissue, and changes in the ratio of maternal to paternal contributions strongly impact on endosperm development. The fertilization independent seed (FIS) Polycomb Repressive Complex 2 (PRC2) is essential for endosperm development; however, the underlying forces that led to the evolution of the FIS-PRC2 remained unknown. Here, we show that the functional requirement of the FIS-PRC2 can be bypassed by increasing the ratio of maternal to paternal genomes in the endosperm, suggesting that the main functional requirement of the FIS-PRC2 is to balance parental genome contributions and to reduce genetic conflict. We furthermore reveal that the AGAMOUS LIKE (AGL) gene AGL62 acts as a dosage-sensitive seed size regulator and that reduced expression of AGL62 might be responsible for reduced size of seeds with increased maternal genome dosage.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
FERTILIZATION-INDEPENDENT SEED-Polycomb Repressive Complex 2 Plays a Dual Role in Regulating Type I MADS-Box Genes in Early Endosperm Development.Plant Physiol. 2018 May;177(1):285-299. doi: 10.1104/pp.17.00534. Epub 2018 Mar 9. Plant Physiol. 2018. PMID: 29523711 Free PMC article.
-
Genome-wide transcript profiling of endosperm without paternal contribution identifies parent-of-origin-dependent regulation of AGAMOUS-LIKE36.PLoS Genet. 2011 Feb;7(2):e1001303. doi: 10.1371/journal.pgen.1001303. Epub 2011 Feb 17. PLoS Genet. 2011. PMID: 21379330 Free PMC article.
-
Two MADS-box proteins, AGL9 and AGL15, recruit the FIS-PRC2 complex to trigger the phase transition from endosperm proliferation to embryo development in Arabidopsis.Mol Plant. 2024 Jul 1;17(7):1110-1128. doi: 10.1016/j.molp.2024.05.011. Epub 2024 Jun 1. Mol Plant. 2024. PMID: 38825830
-
Epigenetic mechanisms governing seed development in plants.EMBO Rep. 2006 Dec;7(12):1223-7. doi: 10.1038/sj.embor.7400854. EMBO Rep. 2006. PMID: 17139298 Free PMC article. Review.
-
[Genomic imprinting and seed development].Yi Chuan. 2005 Jul;27(4):665-70. Yi Chuan. 2005. PMID: 16120596 Review. Chinese.
Cited by
-
Cross-Talk Between Sporophyte and Gametophyte Generations Is Promoted by CHD3 Chromatin Remodelers in Arabidopsis thaliana.Genetics. 2016 Jun;203(2):817-29. doi: 10.1534/genetics.115.180141. Epub 2016 Apr 13. Genetics. 2016. PMID: 27075727 Free PMC article.
-
Plant polymerase IV sensitizes chromatin through histone modifications to preclude spread of silencing into protein-coding domains.Genome Res. 2023 May;33(5):715-728. doi: 10.1101/gr.277353.122. Epub 2023 Jun 5. Genome Res. 2023. PMID: 37277199 Free PMC article.
-
Perturbation of parentally biased gene expression during interspecific hybridization.PLoS One. 2015 Feb 26;10(2):e0117293. doi: 10.1371/journal.pone.0117293. eCollection 2015. PLoS One. 2015. PMID: 25719202 Free PMC article.
-
Disruption of endosperm development is a major cause of hybrid seed inviability between Mimulus guttatus and Mimulus nudatus.New Phytol. 2016 May;210(3):1107-20. doi: 10.1111/nph.13842. Epub 2016 Jan 29. New Phytol. 2016. PMID: 26824345 Free PMC article.
-
Evidence for maternal control of seed size in maize from phenotypic and transcriptional analysis.J Exp Bot. 2016 Mar;67(6):1907-17. doi: 10.1093/jxb/erw006. Epub 2016 Jan 29. J Exp Bot. 2016. PMID: 26826570 Free PMC article.
References
-
- Drews GN, Yadegari R (2002) Development and function of the angiosperm female gametophyte. Annu Rev Genet 36: 99–124. - PubMed
-
- Ingram GC (2010) Family life at close quarters: communication and constraint in angiosperm seed development. Protoplasma 247: 195–214. - PubMed
-
- Costa LM, Gutierrez-Marcos JF, Dickinson HG (2004) More than a yolk: the short life and complex times of the plant endosperm. Trends Plant Sci 9: 507–514. - PubMed
-
- Brown RC, Lemmon BE, Nguyen H, Olsen O-A (1999) Development of the endosperm in Arabidopsis thaliana. Sex Plant Reprod 12: 32–42.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

