Mutational spectrum drives the rise of mutator bacteria
- PMID: 23326242
- PMCID: PMC3542065
- DOI: 10.1371/journal.pgen.1003167
Mutational spectrum drives the rise of mutator bacteria
Abstract
Understanding how mutator strains emerge in bacterial populations is relevant both to evolutionary theory and to reduce the threat they pose in clinical settings. The rise of mutator alleles is understood as a result of their hitchhiking with linked beneficial mutations, although the factors that govern this process remain unclear. A prominent but underappreciated fact is that each mutator allele increases only a specific spectrum of mutational changes. This spectrum has been speculated to alter the distribution of fitness effects of beneficial mutations, potentially affecting hitchhiking. To study this possibility, we analyzed the fitness distribution of beneficial mutations generated from different mutator and wild-type Escherichia coli strains. Using antibiotic resistance as a model system, we show that mutational spectra can alter these distributions substantially, ultimately determining the competitive ability of each strain across environments. Computer simulation showed that the effect of mutational spectrum on hitchhiking dynamics follows a non-linear function, implying that even slight spectrum-dependent fitness differences are sufficient to alter mutator success frequency by several orders of magnitude. These results indicate an unanticipated central role for the mutational spectrum in the evolution of bacterial mutation rates. At a practical level, this study indicates that knowledge of the molecular details of resistance determinants is crucial for minimizing mutator evolution during antibiotic therapy.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
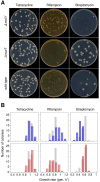
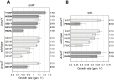
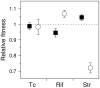
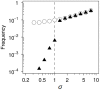
Similar articles
-
Fitness evolution and the rise of mutator alleles in experimental Escherichia coli populations.Genetics. 2002 Oct;162(2):557-66. doi: 10.1093/genetics/162.2.557. Genetics. 2002. PMID: 12399371 Free PMC article.
-
Experimental analysis of molecular events during mutational periodic selections in bacterial evolution.Genetics. 2000 Dec;156(4):1493-501. doi: 10.1093/genetics/156.4.1493. Genetics. 2000. PMID: 11102352 Free PMC article.
-
Bypass of genetic constraints during mutator evolution to antibiotic resistance.Proc Biol Sci. 2015 Apr 7;282(1804):20142698. doi: 10.1098/rspb.2014.2698. Proc Biol Sci. 2015. PMID: 25716795 Free PMC article.
-
Experimental evolution and the dynamics of genomic mutation rate modifiers.Heredity (Edinb). 2014 Nov;113(5):375-80. doi: 10.1038/hdy.2014.49. Epub 2014 May 21. Heredity (Edinb). 2014. PMID: 24849169 Free PMC article. Review.
-
Evolution of mutation rates in bacteria.Mol Microbiol. 2006 May;60(4):820-7. doi: 10.1111/j.1365-2958.2006.05150.x. Mol Microbiol. 2006. PMID: 16677295 Review.
Cited by
-
Genome-Wide Biases in the Rate and Molecular Spectrum of Spontaneous Mutations in Vibrio cholerae and Vibrio fischeri.Mol Biol Evol. 2017 Jan;34(1):93-109. doi: 10.1093/molbev/msw224. Epub 2016 Oct 15. Mol Biol Evol. 2017. PMID: 27744412 Free PMC article.
-
Mutation tendency of mutator Plasmodium berghei with proofreading-deficient DNA polymerase δ.Sci Rep. 2016 Nov 15;6:36971. doi: 10.1038/srep36971. Sci Rep. 2016. PMID: 27845384 Free PMC article.
-
Mutation bias and the predictability of evolution.Philos Trans R Soc Lond B Biol Sci. 2023 May 22;378(1877):20220055. doi: 10.1098/rstb.2022.0055. Epub 2023 Apr 3. Philos Trans R Soc Lond B Biol Sci. 2023. PMID: 37004719 Free PMC article. Review.
-
Mutation bias alters the distribution of fitness effects of mutations.PLoS Biol. 2025 Jul 14;23(7):e3003282. doi: 10.1371/journal.pbio.3003282. eCollection 2025 Jul. PLoS Biol. 2025. PMID: 40658723 Free PMC article.
-
Hypermutator emergence in experimental Escherichia coli populations is stress-type dependent.Evol Lett. 2023 May 8;7(4):252-261. doi: 10.1093/evlett/qrad019. eCollection 2023 Aug. Evol Lett. 2023. PMID: 37475751 Free PMC article.
References
-
- Boe L, Danielsen M, Knudsen S, Petersen JB, Maymann J, et al. (2000) The frequency of mutators in populations of Escherichia coli . Mutat Res 448: 47–55. - PubMed
-
- LeClerc JE, Li B, Payne WL, Cebula TA (1996) High mutation frequencies among Escherichia coli and Salmonella pathogens. Science 274: 1208–1211. - PubMed
-
- Sniegowski PD, Gerrish PJ, Lenski RE (1997) Evolution of high mutation rates in experimental populations of E. coli. Nature 387: 703–705. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

