A population genetic model for the maintenance of R2 retrotransposons in rRNA gene loci
- PMID: 23326244
- PMCID: PMC3542110
- DOI: 10.1371/journal.pgen.1003179
A population genetic model for the maintenance of R2 retrotransposons in rRNA gene loci
Abstract
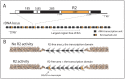
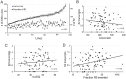
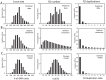
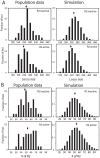
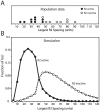
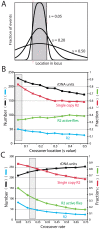
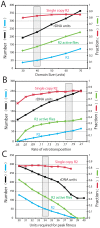
R2 retrotransposable elements exclusively insert into the tandemly repeated rRNA genes, the rDNA loci, of their animal hosts. R2 elements form stable long-term associations with their host, in which all individuals in a population contain many potentially active copies, but only a fraction of these individuals show active R2 retrotransposition. Previous studies have found that R2 RNA transcripts are processed from a 28S co-transcript and that the likelihood of R2-inserted units being transcribed is dependent upon their distribution within the rDNA locus. Here we analyze the rDNA locus and R2 elements from nearly 100 R2-active and R2-inactive individuals from natural populations of Drosophila simulans. Along with previous findings concerning the structure and expression of the rDNA loci, these data were incorporated into computer simulations to model the crossover events that give rise to the concerted evolution of the rRNA genes. The simulations that best reproduce the population data assume that only about 40 rDNA units out of the over 200 total units are actively transcribed and that these transcribed units are clustered in a single region of the locus. In the model, the host establishes this transcription domain at each generation in the region with the fewest R2 insertions. Only if the host cannot avoid R2 insertions within this 40-unit domain are R2 elements active in that generation. The simulations also require that most crossover events in the locus occur in the transcription domain in order to explain the empirical observation that R2 elements are seldom duplicated by crossover events. Thus the key to the long-term stability of R2 elements is the stochastic nature of the crossover events within the rDNA locus, and the inevitable expansions and contractions that introduce and remove R2-inserted units from the transcriptionally active domain.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- Conconi A, Widmer RM, Koller T, Sogo JM (1989) Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle. Cell 57: 753–761. - PubMed
-
- Eickbush TH (2002) R2 and Related Site-specific non-LTR Retrotransposons. pp. 813–835. In Mobile DNA II, N. Craig, R. Craigie, M. Gellert, and A Lambowitz, eds. American Society of Microbiology Press. Washington D.C.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

