A host transcriptional signature for presymptomatic detection of infection in humans exposed to influenza H1N1 or H3N2
- PMID: 23326326
- PMCID: PMC3541408
- DOI: 10.1371/journal.pone.0052198
A host transcriptional signature for presymptomatic detection of infection in humans exposed to influenza H1N1 or H3N2
Abstract
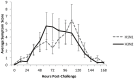
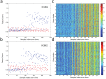
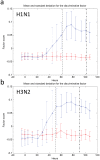
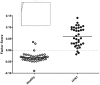
There is great potential for host-based gene expression analysis to impact the early diagnosis of infectious diseases. In particular, the influenza pandemic of 2009 highlighted the challenges and limitations of traditional pathogen-based testing for suspected upper respiratory viral infection. We inoculated human volunteers with either influenza A (A/Brisbane/59/2007 (H1N1) or A/Wisconsin/67/2005 (H3N2)), and assayed the peripheral blood transcriptome every 8 hours for 7 days. Of 41 inoculated volunteers, 18 (44%) developed symptomatic infection. Using unbiased sparse latent factor regression analysis, we generated a gene signature (or factor) for symptomatic influenza capable of detecting 94% of infected cases. This gene signature is detectable as early as 29 hours post-exposure and achieves maximal accuracy on average 43 hours (p = 0.003, H1N1) and 38 hours (p-value = 0.005, H3N2) before peak clinical symptoms. In order to test the relevance of these findings in naturally acquired disease, a composite influenza A signature built from these challenge studies was applied to Emergency Department patients where it discriminates between swine-origin influenza A/H1N1 (2009) infected and non-infected individuals with 92% accuracy. The host genomic response to Influenza infection is robust and may provide the means for detection before typical clinical symptoms are apparent.
Conflict of interest statement
Figures
References
-
- Lambert SB, Whiley DM, O'Neill NT, Andrews EC, Canavan FM, et al. (2008) Comparing nose-throat swabs and nasopharyngeal aspirates collected from children with symptoms for respiratory virus identification using real-time polymerase chain reaction. Pediatrics 122: e615–620. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

