Nuclear legumain activity in colorectal cancer
- PMID: 23326369
- PMCID: PMC3542341
- DOI: 10.1371/journal.pone.0052980
Nuclear legumain activity in colorectal cancer
Erratum in
- PLoS One. 2013;8(9). doi:10.1371/annotation/05c95441-890f-4707-a1bc-c4d386561191
Abstract
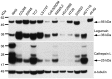
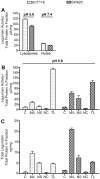
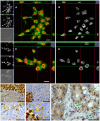
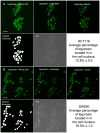
The cysteine protease legumain is involved in several biological and pathological processes, and the protease has been found over-expressed and associated with an invasive and metastatic phenotype in a number of solid tumors. Consequently, legumain has been proposed as a prognostic marker for certain cancers, and a potential therapeutic target. Nevertheless, details on how legumain advances malignant progression along with regulation of its proteolytic activity are unclear. In the present work, legumain expression was examined in colorectal cancer cell lines. Substantial differences in amounts of pro- and active legumain forms, along with distinct intracellular distribution patterns, were observed in HCT116 and SW620 cells and corresponding subcutaneous xenografts. Legumain is thought to be located and processed towards its active form primarily in the endo-lysosomes; however, the subcellular distribution remains largely unexplored. By analyzing subcellular fractions, a proteolytically active form of legumain was found in the nucleus of both cell lines, in addition to the canonical endo-lysosomal residency. In situ analyses of legumain expression and activity confirmed the endo-lysosomal and nuclear localizations in cultured cells and, importantly, also in sections from xenografts and biopsies from colorectal cancer patients. In the HCT116 and SW620 cell lines nuclear legumain was found to make up approximately 13% and 17% of the total legumain, respectively. In similarity with previous studies on nuclear variants of related cysteine proteases, legumain was shown to process histone H3.1. The discovery of nuclear localized legumain launches an entirely novel arena of legumain biology and functions in cancer.
Conflict of interest statement
Figures


Similar articles
-
Nuclear cathepsin L activity is required for cell cycle progression of colorectal carcinoma cells.Biochimie. 2016 Mar;122:208-18. doi: 10.1016/j.biochi.2015.09.003. Epub 2015 Sep 3. Biochimie. 2016. PMID: 26343556
-
Proteolysis mediated by cysteine cathepsins and legumain-recent advances and cell biological challenges.Protoplasma. 2015 May;252(3):755-74. doi: 10.1007/s00709-014-0730-0. Epub 2014 Nov 16. Protoplasma. 2015. PMID: 25398648 Review.
-
Legumain expression in relation to clinicopathologic and biological variables in colorectal cancer.Clin Cancer Res. 2005 Mar 15;11(6):2293-9. doi: 10.1158/1078-0432.CCR-04-1642. Clin Cancer Res. 2005. PMID: 15788679
-
High expression of the cysteine proteinase legumain in colorectal cancer - implications for therapeutic targeting.Eur J Cancer. 2015 Jan;51(1):9-17. doi: 10.1016/j.ejca.2014.10.020. Epub 2014 Nov 11. Eur J Cancer. 2015. PMID: 25466510
-
Mammalian legumain - A lysosomal cysteine protease with extracellular functions?Biochimie. 2019 Nov;166:77-83. doi: 10.1016/j.biochi.2019.06.002. Epub 2019 Jun 7. Biochimie. 2019. PMID: 31181234 Review.
Cited by
-
Structural and functional analysis of cystatin E reveals enzymologically relevant dimer and amyloid fibril states.J Biol Chem. 2018 Aug 24;293(34):13151-13165. doi: 10.1074/jbc.RA118.002154. Epub 2018 Jul 2. J Biol Chem. 2018. PMID: 29967063 Free PMC article.
-
Simvastatin inhibits glucose metabolism and legumain activity in human myotubes.PLoS One. 2014 Jan 8;9(1):e85721. doi: 10.1371/journal.pone.0085721. eCollection 2014. PLoS One. 2014. PMID: 24416446 Free PMC article.
-
Asparagine endopeptidase cleaves apolipoprotein A1 and accelerates pathogenesis of atherosclerosis.J Clin Invest. 2025 May 15;135(10):e185128. doi: 10.1172/JCI185128. eCollection 2025 May 15. J Clin Invest. 2025. PMID: 40371638 Free PMC article.
-
Fabrication of chitosan based nanocomposite with legumain sensitive properties using charge driven self-assembly strategy.J Mater Sci Mater Med. 2018 Aug 18;29(9):142. doi: 10.1007/s10856-018-6149-y. J Mater Sci Mater Med. 2018. PMID: 30121849
-
Asparagine endopeptidase regulates lysosome homeostasis via modulating endomembrane phosphoinositide composition.Cell Death Dis. 2025 Jan 2;15(12):883. doi: 10.1038/s41419-024-07187-3. Cell Death Dis. 2025. PMID: 39743643 Free PMC article.
References
-
- Kembhavi AA, Buttle DJ, Knight CG, Barrett AJ (1993) The Two Cysteine Endopeptidases of Legume Seeds: Purification and Characterization by Use of Specific Fluorometric Assays. Arch Biochem Biophys 303: 208–213. - PubMed
-
- Chen JM, Dando PM, Rawlings ND, Brown MA, Young NE, et al. (1997) Cloning, isolation, and characterization of mammalian legumain, an asparaginyl endopeptidase. J Biol Chem 272: 8090–8098. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

