Structural insight into DFMO resistant ornithine decarboxylase from Entamoeba histolytica: an inkling to adaptive evolution
- PMID: 23326423
- PMCID: PMC3543441
- DOI: 10.1371/journal.pone.0053397
Structural insight into DFMO resistant ornithine decarboxylase from Entamoeba histolytica: an inkling to adaptive evolution
Abstract
Background: Polyamine biosynthetic pathway is a validated therapeutic target for large number of infectious diseases including cancer, giardiasis and African sleeping sickness, etc. α-Difluoromethylornithine (DFMO), a potent drug used for the treatment of African sleeping sickness is an irreversible inhibitor of ornithine decarboxylase (ODC), the first rate limiting enzyme of polyamine biosynthesis. The enzyme ODC of E. histolytica (EhODC) has been reported to exhibit resistance towards DFMO.
Methodology/principal finding: The basis for insensitivity towards DFMO was investigated by structural analysis of EhODC and conformational modifications at the active site. Here, we report cloning, purification and crystal structure determination of C-terminal truncated Entamoeba histolytica ornithine decarboxylase (EhODCΔ15). Structure was determined by molecular replacement method and refined to 2.8 Å resolution. The orthorhombic crystal exhibits P2(1)2(1)2(1) symmetry with unit cell parameters a = 76.66, b = 119.28, c = 179.28 Å. Functional as well as evolutionary relations of EhODC with other ODC homologs were predicted on the basis of sequence analysis, phylogeny and structure.
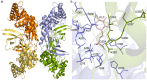
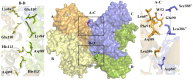
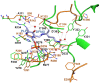
Conclusions/significance: We determined the tetrameric crystal structure of EhODCΔ15, which exists as a dimer in solution. Insensitivity towards DFMO is due to substitution of key substrate binding residues in active site pocket. Additionally, a few more substitutions similar to antizyme inhibitor (AZI), a non-functional homologue of ODCs, were identified in the active site. Here, we establish the fact that EhODC sequence has conserved PLP binding residues; in contrast few substrate binding residues are mutated similar to AZI. Further sequence analysis and structural studies revealed that EhODC may represent as an evolutionary bridge between active decarboxylase and inactive AZI.
Conflict of interest statement
Figures



Similar articles
-
Characterization of the Entamoeba histolytica ornithine decarboxylase-like enzyme.PLoS Negl Trop Dis. 2008 Jan 2;2(1):e115. doi: 10.1371/journal.pntd.0000115. PLoS Negl Trop Dis. 2008. PMID: 18235846 Free PMC article.
-
X-ray structure of ornithine decarboxylase from Trypanosoma brucei: the native structure and the structure in complex with alpha-difluoromethylornithine.Biochemistry. 1999 Nov 16;38(46):15174-84. doi: 10.1021/bi9915115. Biochemistry. 1999. PMID: 10563800
-
Biochemical, mutational and in silico structural evidence for a functional dimeric form of the ornithine decarboxylase from Entamoeba histolytica.PLoS Negl Trop Dis. 2012;6(2):e1559. doi: 10.1371/journal.pntd.0001559. Epub 2012 Feb 28. PLoS Negl Trop Dis. 2012. PMID: 22389745 Free PMC article.
-
Alpha-Difluoromethylornithine, an Irreversible Inhibitor of Polyamine Biosynthesis, as a Therapeutic Strategy against Hyperproliferative and Infectious Diseases.Med Sci (Basel). 2018 Feb 8;6(1):12. doi: 10.3390/medsci6010012. Med Sci (Basel). 2018. PMID: 29419804 Free PMC article. Review.
-
Ornithine decarboxylase as an enzyme target for therapy.Pharmacol Ther. 1992;54(2):195-215. doi: 10.1016/0163-7258(92)90032-u. Pharmacol Ther. 1992. PMID: 1438532 Review.
Cited by
-
Synthesis and Deployment of an Elusive Fluorovinyl Cation Equivalent: Access to Quaternary α-(1'-Fluoro)vinyl Amino Acids as Potential PLP Enzyme Inactivators.J Am Chem Soc. 2017 Oct 11;139(40):14077-14089. doi: 10.1021/jacs.7b04690. Epub 2017 Sep 28. J Am Chem Soc. 2017. PMID: 28906111 Free PMC article.
-
Revisiting Drug Development Against the Neglected Tropical Disease, Amebiasis.Front Cell Infect Microbiol. 2021 Feb 24;10:628257. doi: 10.3389/fcimb.2020.628257. eCollection 2020. Front Cell Infect Microbiol. 2021. PMID: 33718258 Free PMC article. Review.
-
Hydroxylamine Analogue of Agmatine: Magic Bullet for Arginine Decarboxylase.Biomolecules. 2020 Mar 6;10(3):406. doi: 10.3390/biom10030406. Biomolecules. 2020. PMID: 32155745 Free PMC article.
-
The First Insight Into the Supramolecular System of D,L-α-Difluoromethylornithine: A New Antiviral Perspective.Front Chem. 2021 May 13;9:679776. doi: 10.3389/fchem.2021.679776. eCollection 2021. Front Chem. 2021. PMID: 34055746 Free PMC article.
References
-
- Rosas-Arreguín P, Arteaga-Nieto P, Reynoso-Orozco R, Villagómez-Castro JC, Sabanero-López M, et al. (2008) Bursera fagaroides, effect of an ethanolic extract on ornithine decarboxylase (ODC) activity in vitro and on the growth of Entamoeba histolytica . Exp Parasitol 119: 398–402. - PubMed
-
- López-Vallejo F, Castillo R, Yépez-Mulia L, Medina-Franco JL (2011) Benzotriazoles and indazoles are scaffolds with biological activity against Entamoeba histolytica . J Biomol Screen 16: 862–868. - PubMed
-
- Heby O, Roberts SC, Ullman B (2003) Polyamine biosynthetic enzymes as drug targets in parasitic protozoa. Biochem Soc Trans 31: 415–419. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

